- 1、[CV] HyperMorph: Amortized Hyperparameter Learning for Image Registration
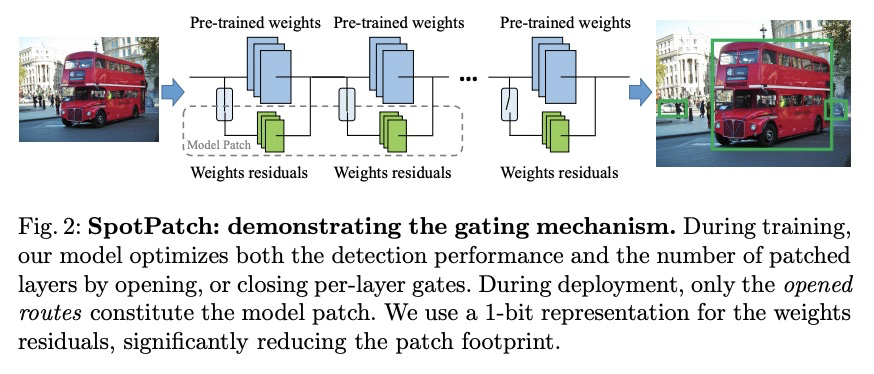
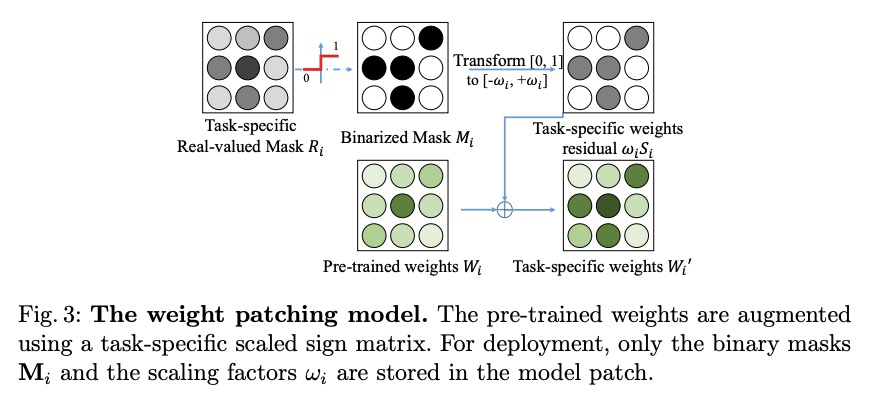
- 2、[CV] SpotPatch: Parameter-Efficient Transfer Learning for Mobile Object Detection
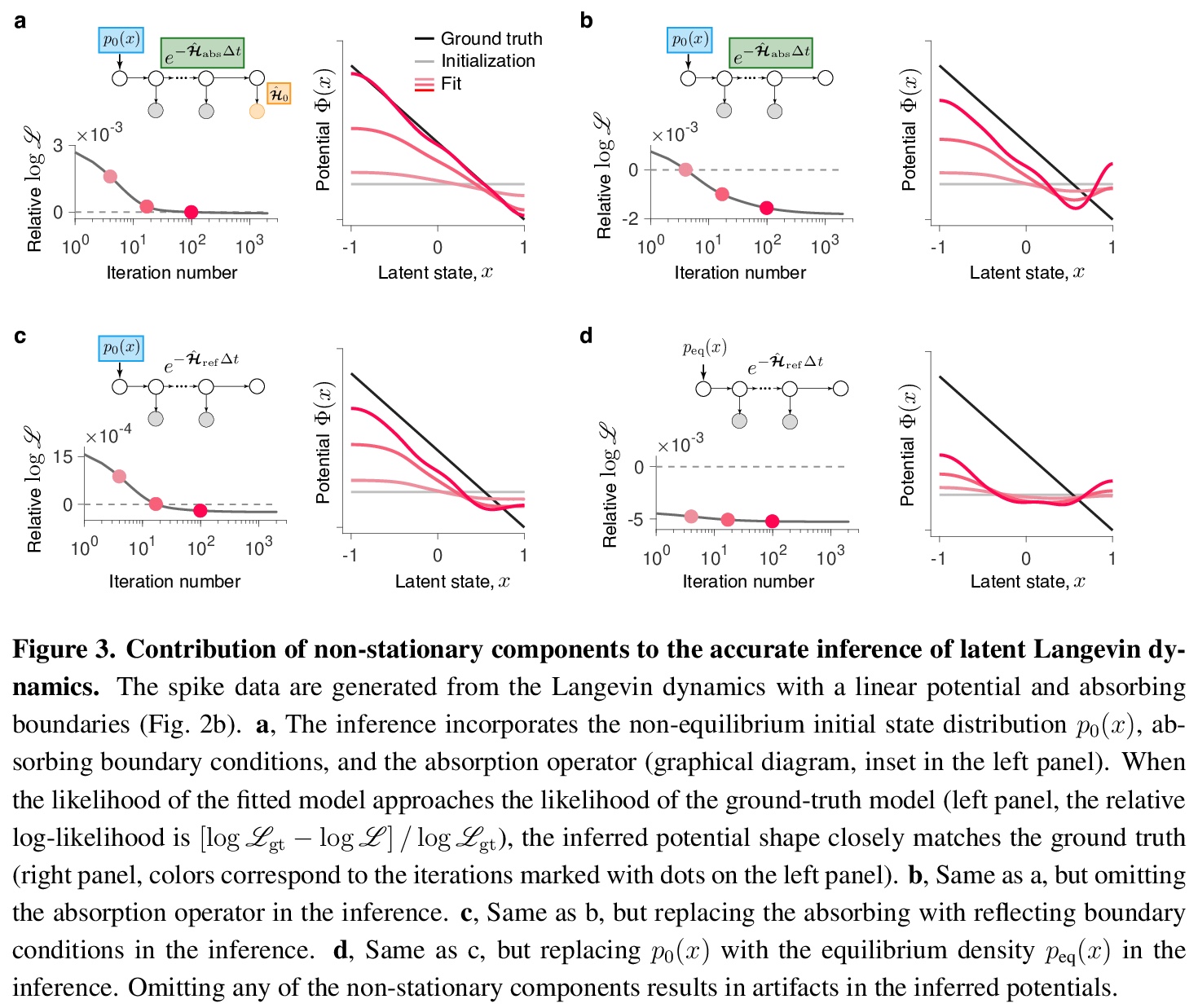
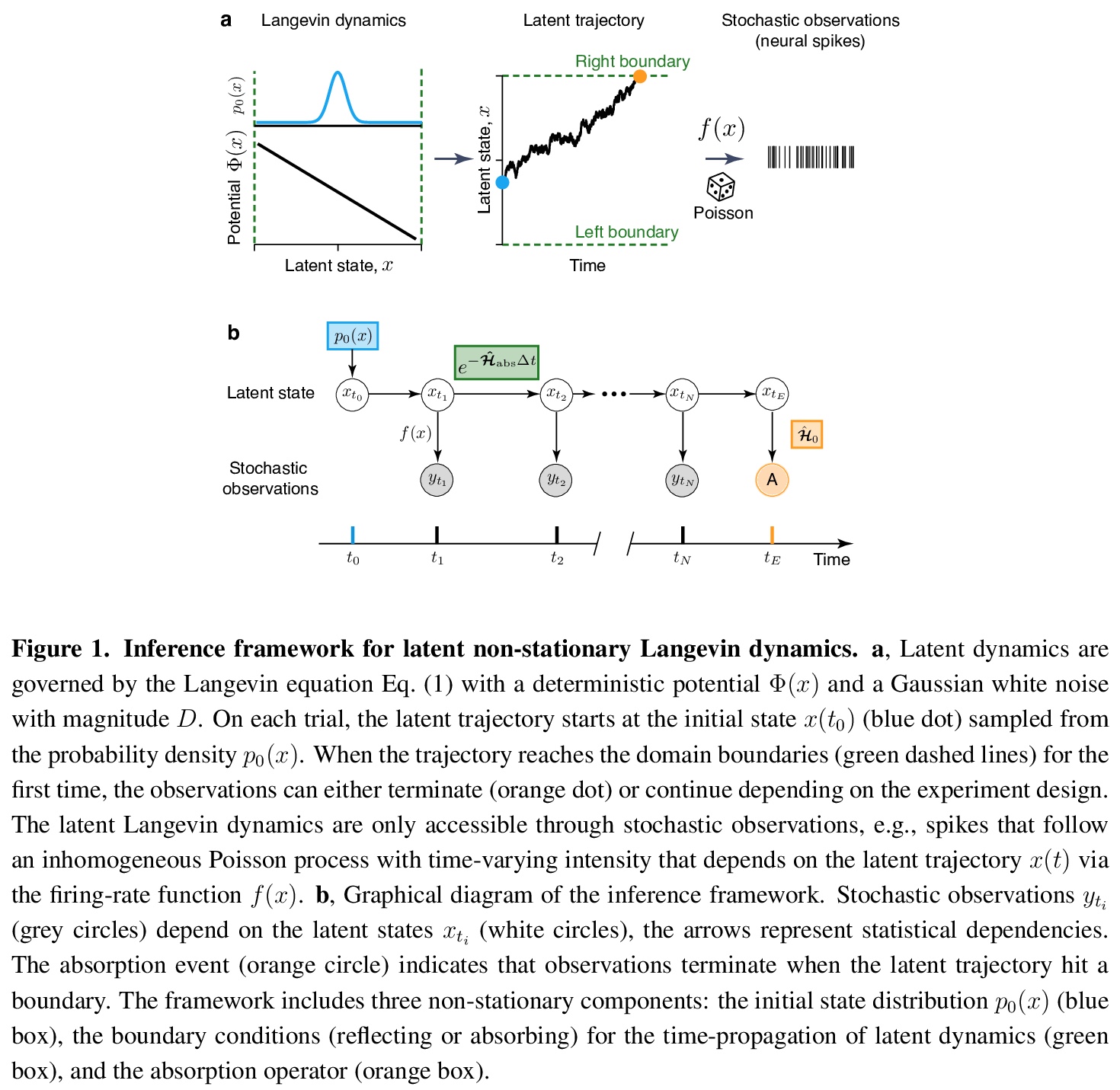
- 3、[LG] Learning non-stationary Langevin dynamics from stochastic observations of latent trajectories
- 4、[LG] Segmentation and genome annotation algorithms
- 5、[CL] I-BERT: Integer-only BERT Quantization
LG - 机器学习 CV - 计算机视觉 CL - 计算与语言 AS - 音频与语音 RO - 机器人 (*表示值得重点关注)
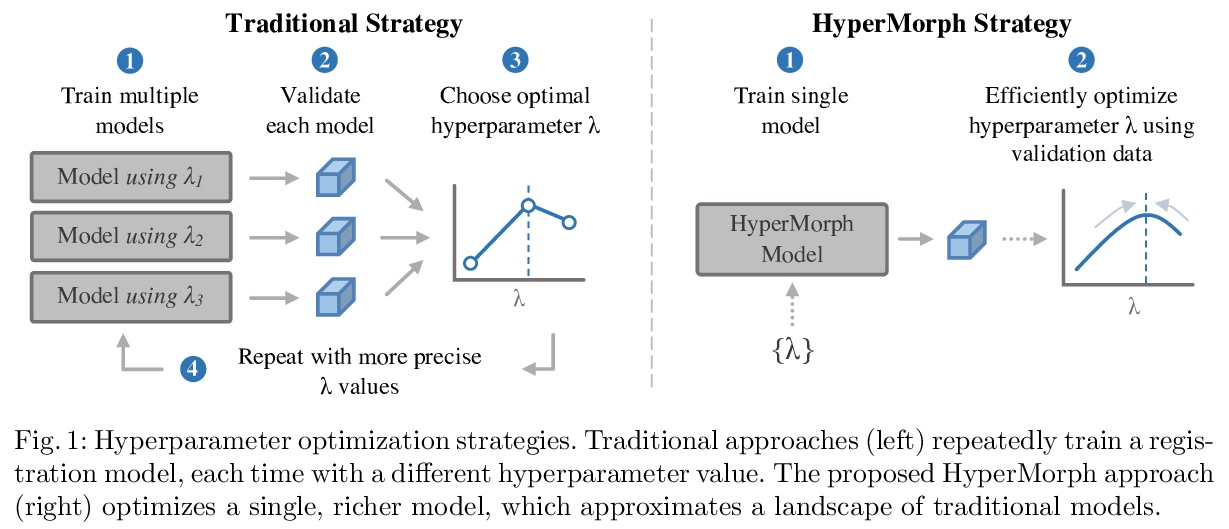
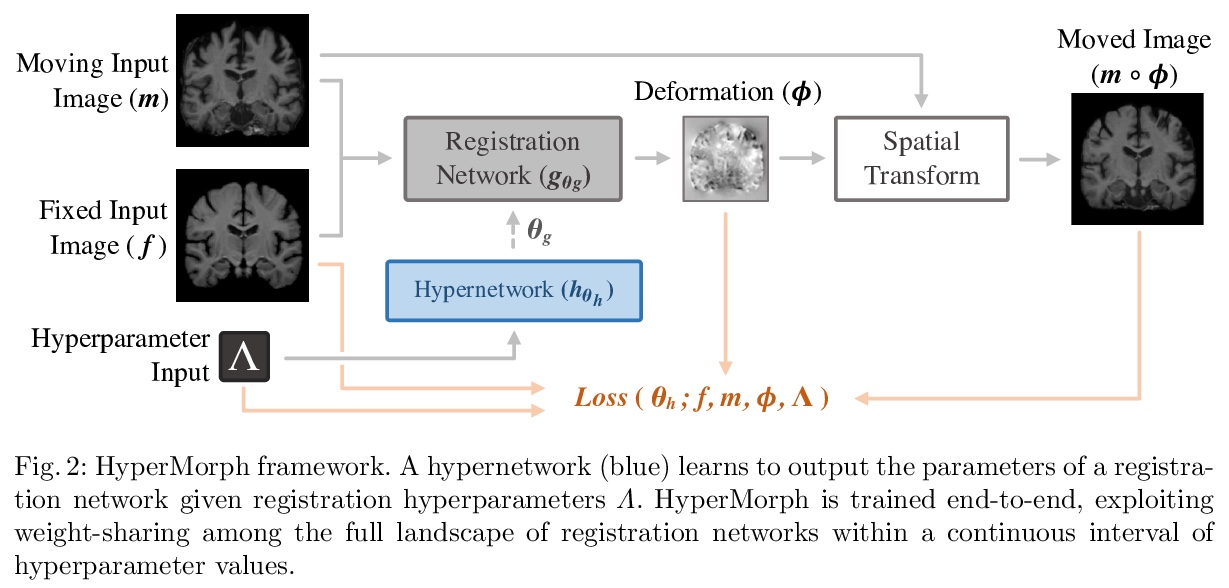
1、[CV] HyperMorph: Amortized Hyperparameter Learning for Image Registration
A Hoopes, M Hoffmann, B Fischl, J Guttag, A V. Dalca
[MGH & MIT]
HyperMorph:摊余超参数学习图像配准。提出基于学习的可变形图像配准策略HyperMorph,消除了训练过程中调整重要配准超参数的需要。该框架学习一个超网络,该网络接收一个输入的超参数,对配准网络进行调制,以产生该超参数值的最优变形场。该策略训练了一个单一的、丰富的模型,可在测试时从一个连续的区间快速、精细地发现超参数值。实验证明,此方法可用来优化多个超参数,比现有搜索策略快得多,减少了计算和人力负担,提高了灵活性。
We present HyperMorph, a learning-based strategy for deformable image registration that removes the need to tune important registration hyperparameters during training. Classical registration methods solve an optimization problem to find a set of spatial correspondences between two images, while learning-based methods leverage a training dataset to learn a function that generates these correspondences. The quality of the results for both types of techniques depends greatly on the choice of hyperparameters. Unfortunately, hyperparameter tuning is time-consuming and typically involves training many separate models with various hyperparameter values, potentially leading to suboptimal results. To address this inefficiency, we introduce amortized hyperparameter learning for image registration, a novel strategy to learn the effects of hyperparameters on deformation fields. The proposed framework learns a hypernetwork that takes in an input hyperparameter and modulates a registration network to produce the optimal deformation field for that hyperparameter value. In effect, this strategy trains a single, rich model that enables rapid, fine-grained discovery of hyperparameter values from a continuous interval at test-time. We demonstrate that this approach can be used to optimize multiple hyperparameters considerably faster than existing search strategies, leading to a reduced computational and human burden and increased flexibility. We also show that this has several important benefits, including increased robustness to initialization and the ability to rapidly identify optimal hyperparameter values specific to a registration task, dataset, or even a single anatomical region - all without retraining the HyperMorph model. Our code is publicly available at > this http URL.
https://weibo.com/1402400261/JC0XuvZnY
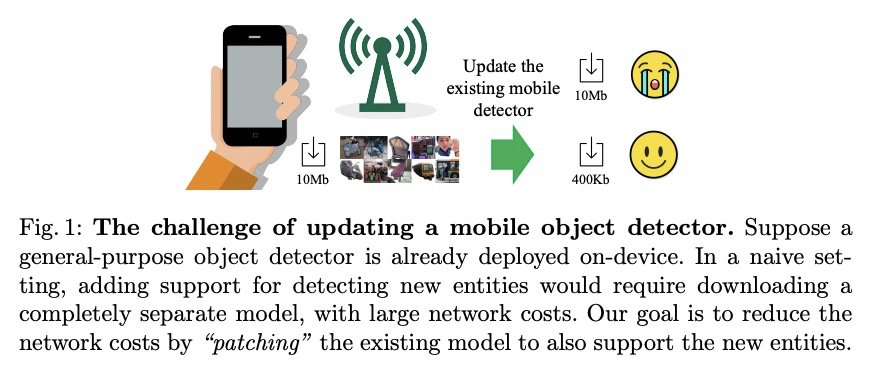
2、[CV] SpotPatch: Parameter-Efficient Transfer Learning for Mobile Object Detection
K Ye, A Kovashka, M Sandler, M Zhu, A Howard, M Fornoni
[University of Pittsburgh & Google Research & DJI Technology LLC]
SpotPatch:面向移动端目标检测的参数高效迁移学习。系统研究了针对目标检测问题的参数高效迁移学习技术,引入了检测全能任务,提出SpotPatch方法,用特定于任务的权重转换和动态路由来最小化学到区块的占用空间。在所有考虑的基准中,SpotPatch提供了与标准权重变换相似的mAP,但参数效率显著提高。
Deep learning based object detectors are commonly deployed on mobile devices to solve a variety of tasks. For maximum accuracy, each detector is usually trained to solve one single specific task, and comes with a completely independent set of parameters. While this guarantees high performance, it is also highly inefficient, as each model has to be separately downloaded and stored. In this paper we address the question: can task-specific detectors be trained and represented as a shared set of weights, plus a very small set of additional weights for each task? The main contributions of this paper are the following: 1) we perform the first systematic study of parameter-efficient transfer learning techniques for object detection problems; 2) we propose a technique to learn a model patch with a size that is dependent on the difficulty of the task to be learned, and validate our approach on 10 different object detection tasks. Our approach achieves similar accuracy as previously proposed approaches, while being significantly more compact.
https://weibo.com/1402400261/JC125opGz
3、[LG] Learning non-stationary Langevin dynamics from stochastic observations of latent trajectories
M Genkin, O Hughes, T A. Engel
[Cold Spring Harbor & University of Michigan]
从潜轨迹随机观测中学习非平稳朗之万动力学。提出一个非参数朗之万方程推断框架,该框架明确模拟了随机观测过程和非平稳潜动力学。该框架考虑了观测系统的非平衡初始状态和最终状态,以及系统动态定义观测持续时间的可能性。忽略这些非稳态成分中的任何一个都会导致不正确的推理,由于数据的非平稳分布,在动态中会产生错误的特征。用神经动力学模型来说明在大脑中作出决定的框架。
Many complex systems operating far from the equilibrium exhibit stochastic dynamics that can be described by a Langevin equation. Inferring Langevin equations from data can reveal how transient dynamics of such systems give rise to their function. However, dynamics are often inaccessible directly and can be only gleaned through a stochastic observation process, which makes the inference challenging. Here we present a non-parametric framework for inferring the Langevin equation, which explicitly models the stochastic observation process and non-stationary latent dynamics. The framework accounts for the non-equilibrium initial and final states of the observed system and for the possibility that the system’s dynamics define the duration of observations. Omitting any of these non-stationary components results in incorrect inference, in which erroneous features arise in the dynamics due to non-stationary data distribution. We illustrate the framework using models of neural dynamics underlying decision making in the brain.
https://weibo.com/1402400261/JC17cr4dh
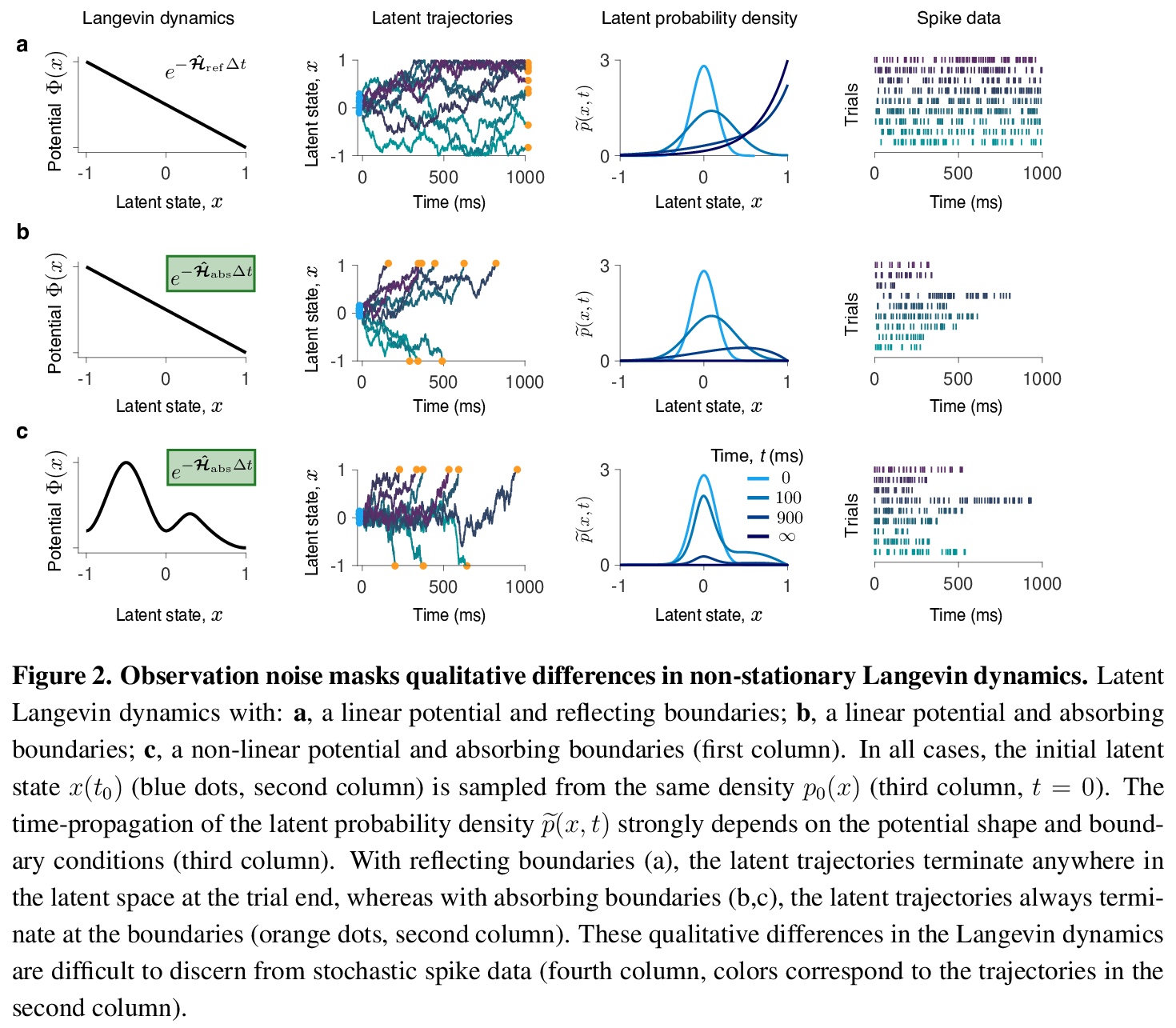
4、[LG] Segmentation and genome annotation algorithms
M W Libbrecht, R C Chan, M M Hoffman
[Simon Fraser University & University of Toronto]
分段和基因组注释算法综述。分段和基因组注释算法(SAGA)被广泛用于了解基因组活动和基因调控。这些算法将表观基因组数据集作为输入,如染色质免疫沉淀测序(ChIP-seq)对组蛋白修饰或转录因子结合的测量。它们对基因组进行分区,并为每个片段分配一个标签,使具有相同标签的位置表现出输入数据的相似模式。SAGA算法发现活动的类别,如启动子、增强子,或部分基因的事先不知道的已知基因组元素。本文回顾了作为这些方法基础的方法论框架,回顾了这一基本框架的变体和改进,对现有的大规模参考标记进行了编目,并讨论了未来工作的前景。
Segmentation and genome annotation (SAGA) algorithms are widely used to understand genome activity and gene regulation. These algorithms take as input epigenomic datasets, such as chromatin immunoprecipitation-sequencing (ChIP-seq) measurements of histone modifications or transcription factor binding. They partition the genome and assign a label to each segment such that positions with the same label exhibit similar patterns of input data. SAGA algorithms discover categories of activity such as promoters, enhancers, or parts of genes without prior knowledge of known genomic elements. In this sense, they generally act in an unsupervised fashion like clustering algorithms, but with the additional simultaneous function of segmenting the genome. Here, we review the common methodological framework that underlies these methods, review variants of and improvements upon this basic framework, catalogue existing large-scale reference annotations, and discuss the outlook for future work.
https://weibo.com/1402400261/JC1cZvJi5
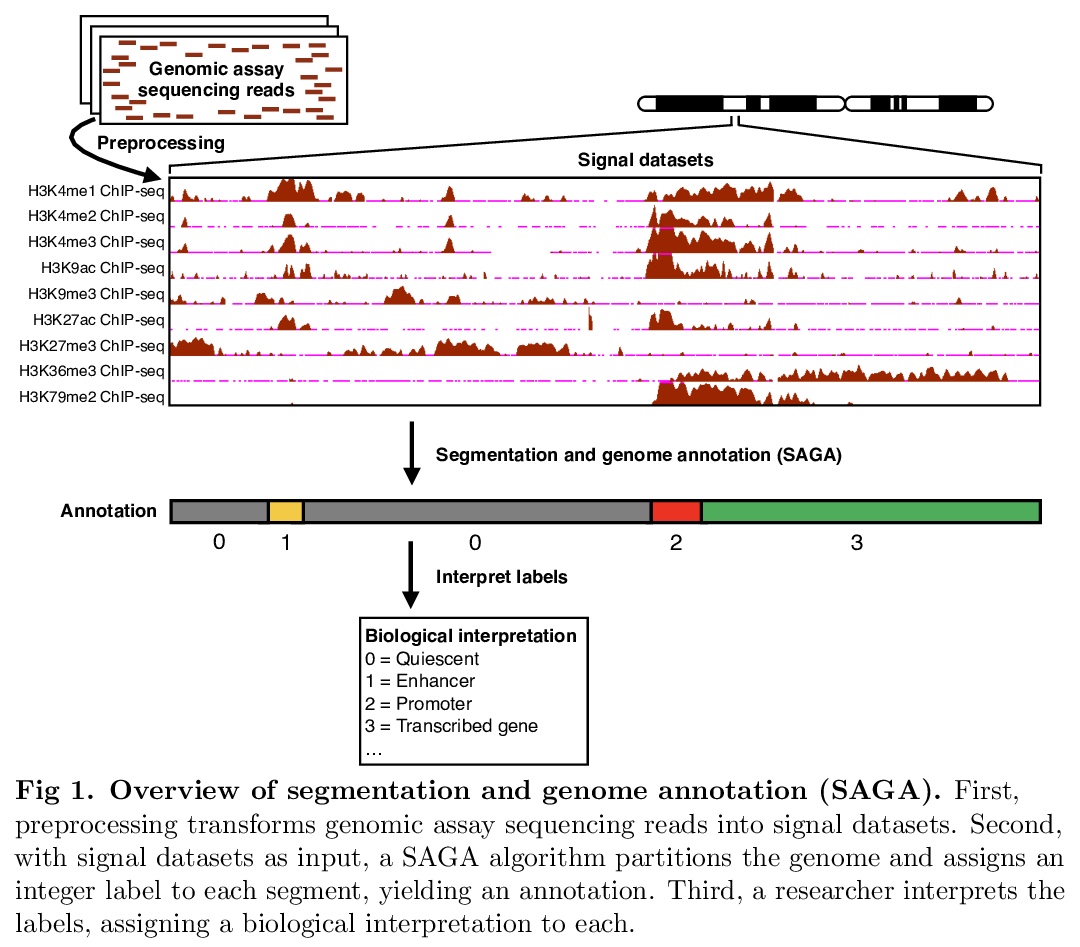
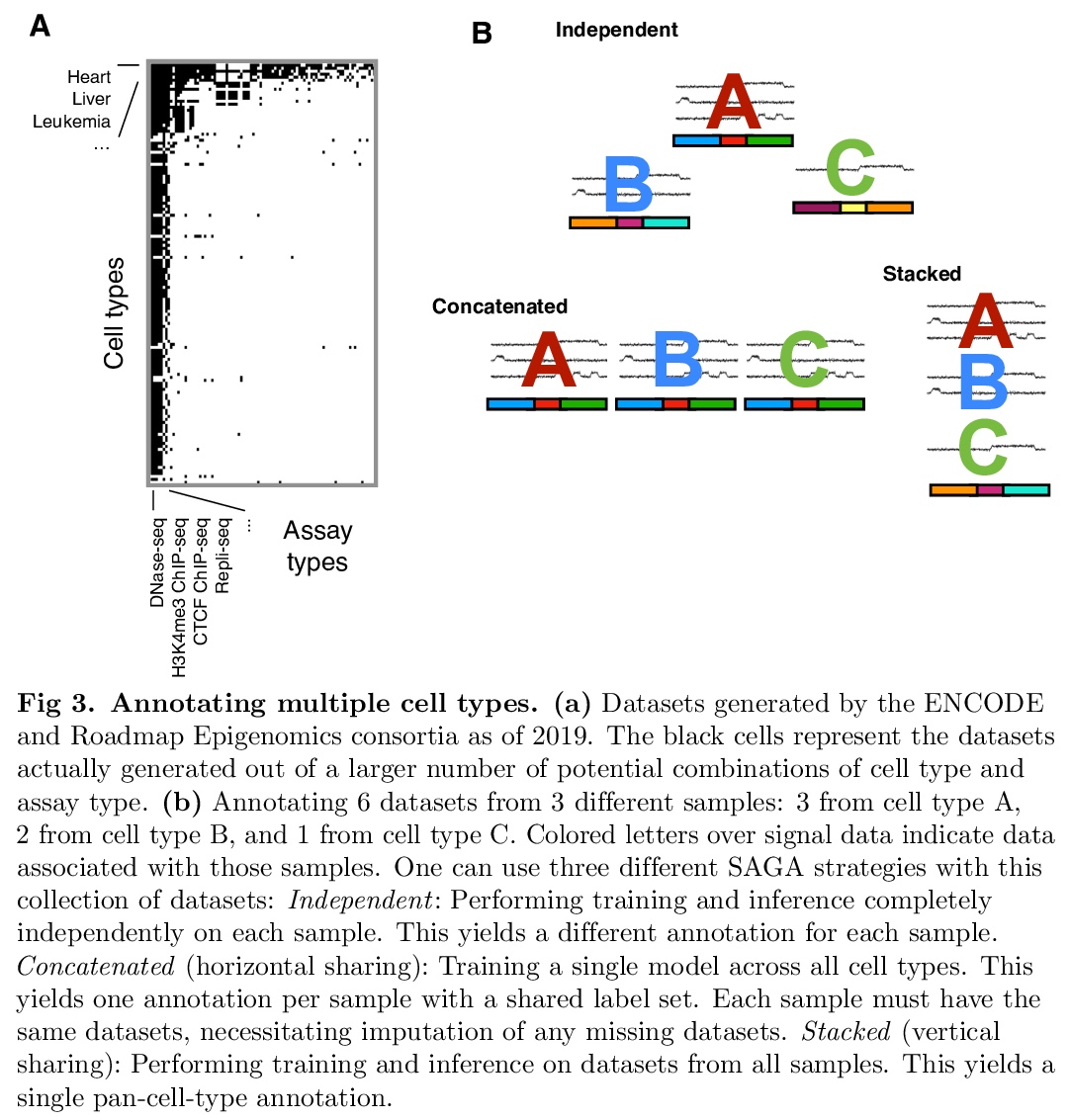
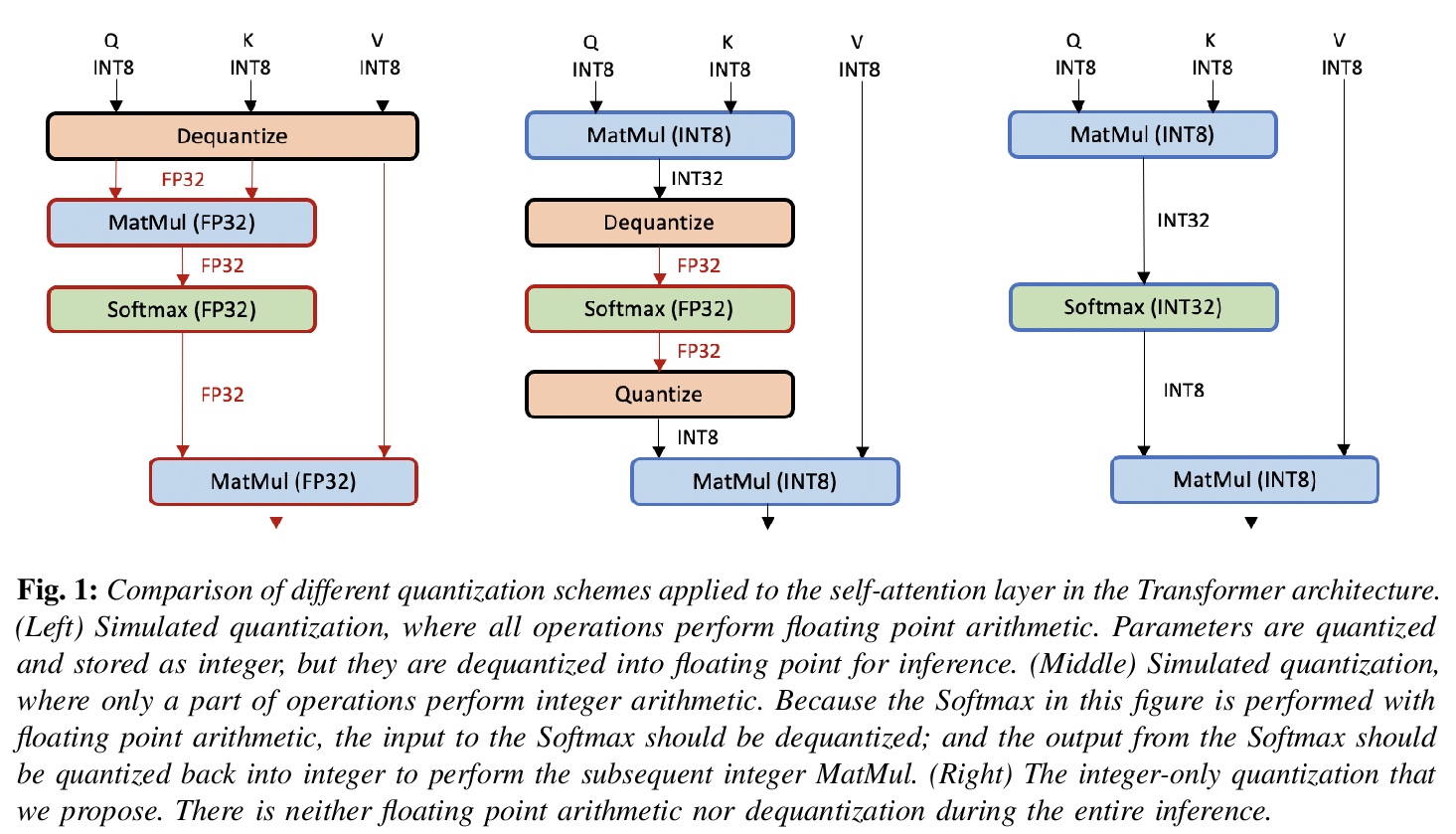
5、[CL] I-BERT: Integer-only BERT Quantization
S Kim, A Gholami, Z Yao, M W. Mahoney, K Keutzer
[UC Berkeley]
I-BERT:纯整数BERT量化。为基于Transformer的模型提出了一种新型的纯整数量化方案,对整个推理过程进行量化。I-BERT的核心是非线性运算如GELU、Softmax和LayerNorm的近似方法,可用整数计算来进行近似。实验表明,在平均GLUE得分方面,该方法比浮点基线模型有所提高。
Transformer based models, like BERT and RoBERTa, have achieved state-of-the-art results in many Natural Language Processing tasks. However, their memory footprint, inference latency, and power consumption are prohibitive for many edge processors, and it has been a challenge to deploy these models for edge applications and devices that have resource constraints. While quantization can be a viable solution to this, previous work on quantizing Transformer based models uses floating-point arithmetic during inference, thus limiting model deployment on many edge processors. In this work, we propose a novel integer-only quantization scheme for Transformer based models that quantizes the entire inference process. In particular, we demonstrate how to approximate nonlinear operations in Transformer architectures, e.g., GELU, Softmax, and Layer Normalization, with lightweight integer computations. We use those approximations in our method, I-BERT, with an end-to-end integer-only inference, and without any floating point calculation. We test our approach on GLUE downstream tasks using RoBERTa-Base and RoBERTa-Large. For both cases, with an 8-bit integer-only quantization scheme, I-BERT achieves similar accuracy as compared to the full-precision baseline.
https://weibo.com/1402400261/JC1gHm2Tz