For similarity analysis of time-series data, Euclidean distance is typically used as a similarity measure. 对于时间序列数据的相似性分析,欧氏距离通常用作相似性度量。
The smaller the distance between two sets of time-series data, the more similar are the two series. 两组时间序列数据之间的距离越小,两个序列越相似。
However, we cannot directly apply the Euclidean distance. Instead, we need to consider differences in the baseline and scale (or amplitude) of our two series. 然而,我们不能直接应用欧几里得距离。相反,我们需要考虑我们两个系列的基线和尺度(或振幅)的差异。·
For example, one stock’s value may have a baseline of around $20 and fluctuate with a relatively large amplitude (such as between $15 and $25), while another could have a baseline of around $100 and fluctuate with a relatively small amplitude (such as between $90 and $110). 例如,一只股票的价值可能有一个20美元左右的基线,波动幅度相对较大(如15美元至25美元之间),而另一只股票的价值可能有一个100美元左右的基线,波动幅度相对较小(如90美元至110美元之间)。
Offset 抵消: the distance from one baseline to another.
Normalisation transformation: one way to solve the baseline and scale problem 标准化转换:解决基线和规模问题的一种方法
A sequence 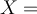 can be replaced by normalised sequence
can be replaced by normalised sequence 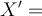 using
using 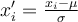 , where
, where 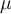 is the mean value of sequence
is the mean value of sequence 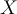 and
and 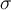 is the standard deviation of
is the standard deviation of 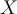 .
.
Sequence matching does not require perfect matching. 序列匹配不需要完美匹配。
We should allow for pairs of subsequences to match if they are of the same shape, but differ due to the presence of gaps within a sequence (where one of the series may be missing some of the values that exist in the other) or differences in offsets or amplitudes. 如果子序列具有相同的形状,但由于序列中存在间隙(其中一个序列可能缺少另一个序列中存在的一些值)或偏移或振幅的差异而不同,我们应该允许子序列对匹配。
“How can subsequence matching be performed to allow for such differences?“
Users or experts can specify parameters such as a sliding window size, the width of an envelope for similarity, the maximum gap, a matching fraction, and so on. 用户或专家可以指定参数,如滑动窗口大小、相似度包络宽度、最大间隙、匹配分数等。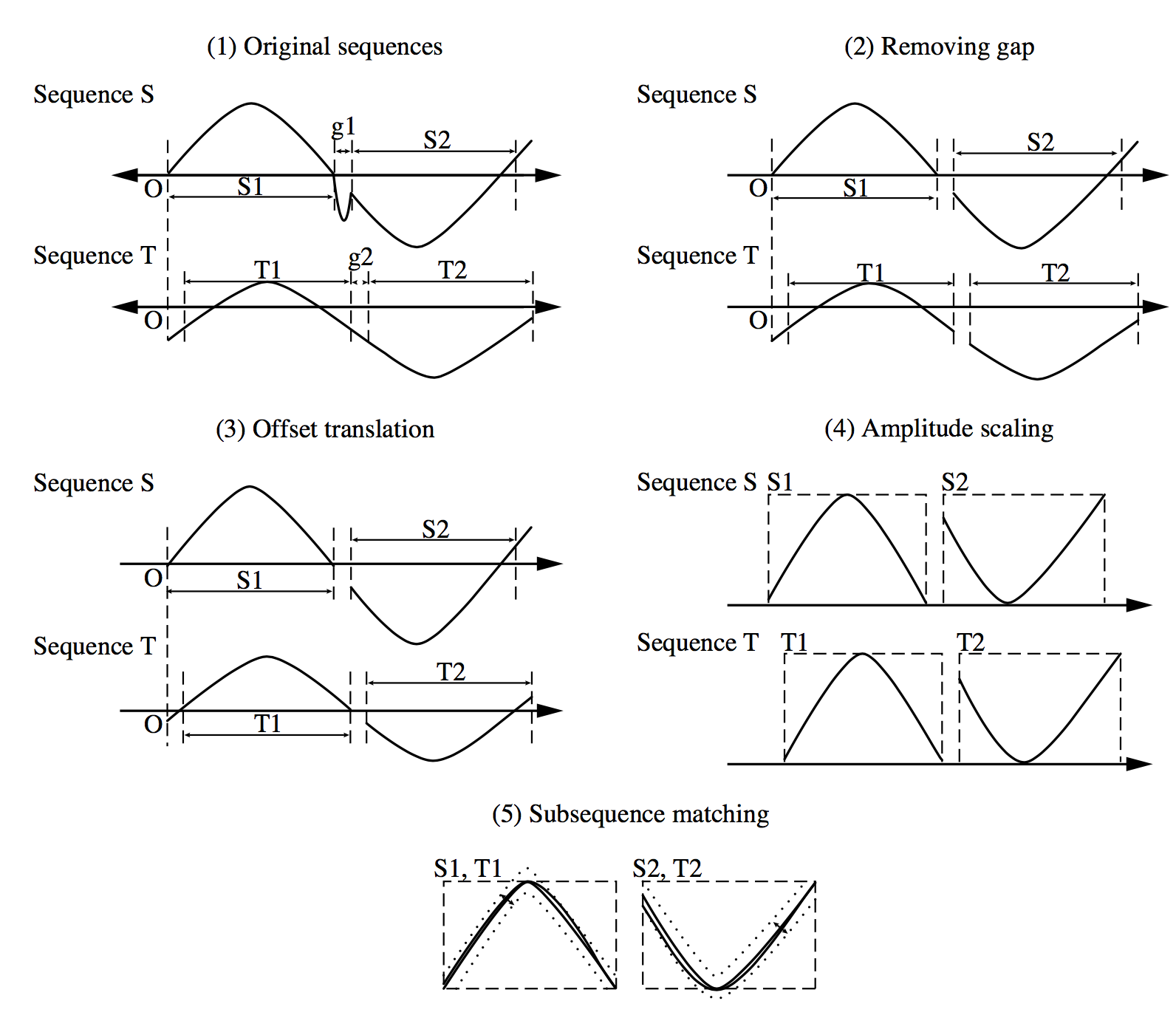
> Illustration of the process for subsequence matching
As illustrated, we begin with two sequences in their original form (1).
(2) gaps are removed.
(3) normalisation with respect to offset translation (where one time series is adjusted to align with the other by shifting the baseline or phase)
(4) amplitude scaling. Two subsequences are considered similar and can be matched if one lies within an envelope of 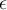 width around the other, ignoring outliers. Two sequences are similar if they have enough nonoverlapping time-ordered pairs of similar subsequences.
width around the other, ignoring outliers. Two sequences are similar if they have enough nonoverlapping time-ordered pairs of similar subsequences.
Based on the above, a similarity search that handles gaps and differences in offsets and amplitudes can be performed by the following steps:
- Atomic matching: Normalise the data. Find all pairs of gap-free windows of a small length that are similar.
- Window stitching: Stitch similar windows to form pairs of large similar subsequences, allowing gaps between atomic matches.
- Subsequence ordering: Linearly order the subsequence matches to determine whether enough similar pieces exist.
With such processing, sequences of similar shape but with gaps or differences in offsets or amplitudes can be found to match each other or to match query templates.

