介绍
EDTA (The Extensive de novo TE Annotator)
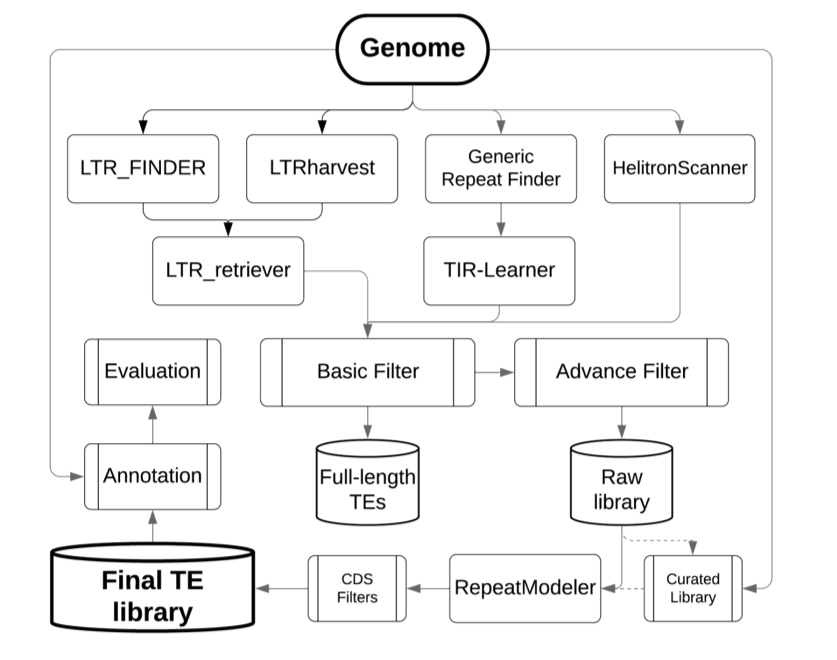
使用
软件下载
cd /home/shwzhao/bin/SingularityDir/SINGULARITY_CACHEDIR=./export SINGULARITY_CACHEDIRsingularity pull EDTA.sif docker://oushujun/edta:2.0.0singularity exec /home/shwzhao/bin/SingularityDir/EDTA.sif EDTA.pl -h## INFO: Converting SIF file to temporary sandbox...#### ########################################################## ##### Extensive de-novo TE Annotator (EDTA) v2.0.0 ###### ##### Shujun Ou (shujun.ou.1@gmail.com) ###### ########################################################## .......
参数说明
具体看参考吧
- EDTA.pl
--curatedlib
--sensitive
--rmout
--exclude
--u
- EDTA_raw.pl
--type
- lib-test.pl
检验注释性能
运行
mkdir testcd testcp /home/train/public_data/genome/Arabidopsis_thaliana/Arabidopsis_thaliana.genome.fa .singularity exec /home/shwzhao/bin/SingularityDir/EDTA.sif EDTA.pl --genome ./Arabidopsis_thaliana.genome.fa --overwrite 0 --sensitive 1 --anno 1 --threads 20
genome=/pfs/proj/nobackup/fs/projnb10/hpc2nstor2024-021/shwzhao/01_research/03_assemble2/6_polish/genome.all.polished.faEDTA.pl \--genome ${genome} \--overwrite 1 \--sensitive 1 \--anno 1 \--threads 50 \1> edta.out \2> edta.errls -lh## total 3.2G## -rw-rw----+ 1 shwzhao ps30521 9.4K Oct 23 10:06 edta.err## -rw-rw----+ 1 shwzhao ps30521 2.0K Oct 23 11:43 edta.out## lrwxrwxrwx 1 shwzhao ps30521 120 Oct 20 11:39 genome.all.polished.fa -> /pfs/proj/nobackup/fs/projnb10/hpc2nstor2024-021/shwzhao/01_research/03_assemble2/6_polish/genome.all.polished.fa## -rw-rw----+ 1 shwzhao ps30521 752M Oct 20 11:39 genome.all.polished.fa.mod## drwxrws---+ 2 shwzhao ps30521 4.0K Oct 23 11:43 genome.all.polished.fa.mod.EDTA.anno## drwxrws---+ 2 shwzhao ps30521 20K Oct 21 00:38 genome.all.polished.fa.mod.EDTA.combine## drwxrws---+ 3 shwzhao ps30521 12K Oct 23 10:06 genome.all.polished.fa.mod.EDTA.final## -rw-rw----+ 1 shwzhao ps30521 3.9M Oct 23 10:06 genome.all.polished.fa.mod.EDTA.intact.gff3## drwxrws---+ 5 shwzhao ps30521 4.0K Oct 21 00:23 genome.all.polished.fa.mod.EDTA.raw## -rw-rw----+ 1 shwzhao ps30521 121M Oct 23 11:43 genome.all.polished.fa.mod.EDTA.TEanno.gff3## -rw-rw----+ 1 shwzhao ps30521 288K Oct 23 11:43 genome.all.polished.fa.mod.EDTA.TEanno.sum## -rw-rw----+ 1 shwzhao ps30521 8.5M Oct 23 10:03 genome.all.polished.fa.mod.EDTA.TElib.fa## -rw-rw----+ 1 shwzhao ps30521 752M Oct 23 11:43 genome.all.polished.fa.mod.MAKER.masked## -rw-rw----+ 1 shwzhao ps30521 440 Oct 20 11:29 run.sh
2.4 可视化
以后补充
grep -v "##" Arabidopsis_thaliana.genome.fa.mod.EDTA.TEanno.gff3 | cut -f 1-5,7 | sed '1i seqid\tsource\tsequence_ontology\tstart\tend\tstrand' > ara.edta.tsv
参考
github 地址:https://github.com/oushujun/EDTA
bilibili | CGM第七十九期 区树俊博士的报告
xuzhougeng | 有了它,基因组重复序列注释一次全搞定

