介绍
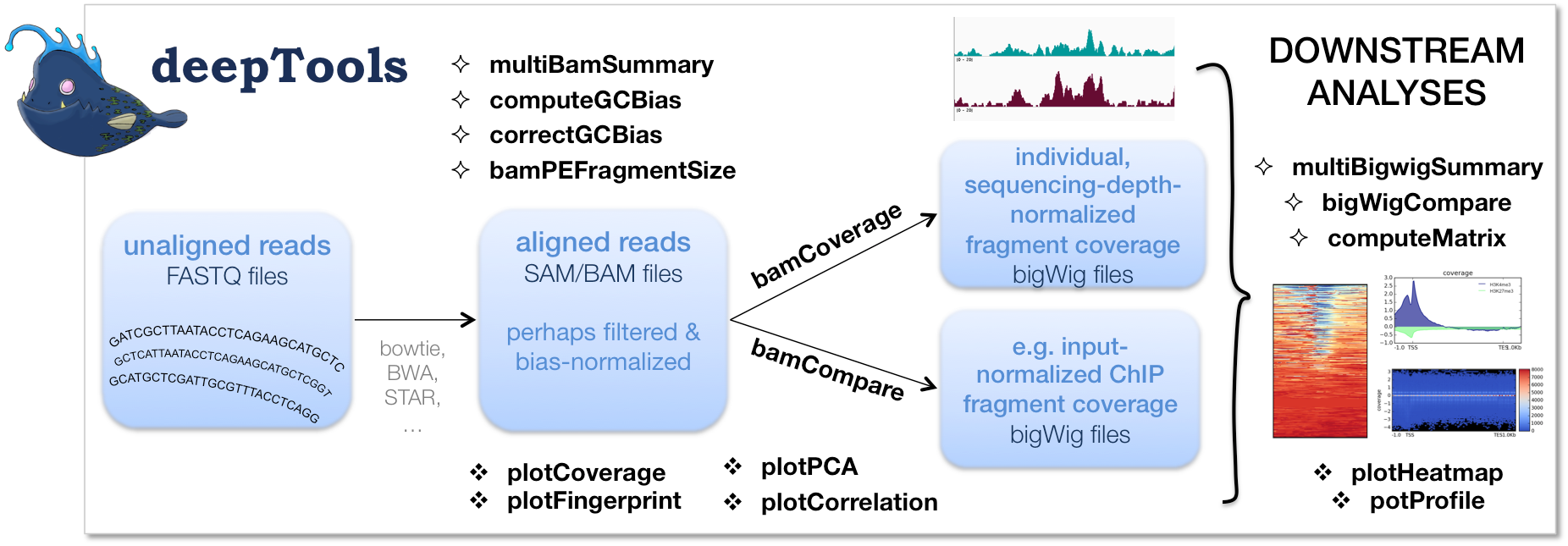
BAM 和 bigWig 文件的处理
multiBamSummary
multiBigwigSummary
correctGCBias
bamCoverage
bamCoverage \ --bam a.bam \ -o a.SeqDepthNorm.bw \ --binSize 10 \ --normalizeUsing RPGC \ --effectiveGenomeSize 2150570000 \ --ignoreForNormalization chrX \ --extendReadsbamCoverage \ --bam myAlignedReads.bam \ --outFileName myCoverageFile.bigWig \ --outFileFormat bigwig \ --fragmentLength 200 \ --ignoreDuplicates \ --scaleFactor 0.5
bamCompare
bigwigCompare
bigwigAverage
computeMatrix
alignmentSieve
参考
- https://github.com/deeptools/deepTools
- https://deeptools.readthedocs.io/en/latest/
- deepTools工具说明


