在本教程中,我们将学习如何通过一些经典的marker基因计算细胞周期阶段评分,并在预处理过程中将其从scRNA-seq数据中进行回归处理,来减轻细胞周期异质性对下游数据分析的影响。
加载所需的R包和数据
library(Seurat)# Read in the expression matrix The first row is a header row, the first column is rownamesexp.mat <- read.table(file = "../data/nestorawa_forcellcycle_expressionMatrix.txt", header = TRUE,as.is = TRUE, row.names = 1)# A list of cell cycle markers, from Tirosh et al, 2015, is loaded with Seurat. We can segregate this list into markers of G2/M phase and markers of S phase# 查看细胞周期marker基因cc.genes$s.genes[1] "MCM5" "PCNA" "TYMS" "FEN1" "MCM2" "MCM4" "RRM1"[8] "UNG" "GINS2" "MCM6" "CDCA7" "DTL" "PRIM1" "UHRF1"[15] "MLF1IP" "HELLS" "RFC2" "RPA2" "NASP" "RAD51AP1" "GMNN"[22] "WDR76" "SLBP" "CCNE2" "UBR7" "POLD3" "MSH2" "ATAD2"[29] "RAD51" "RRM2" "CDC45" "CDC6" "EXO1" "TIPIN" "DSCC1"[36] "BLM" "CASP8AP2" "USP1" "CLSPN" "POLA1" "CHAF1B" "BRIP1"[43] "E2F8"$g2m.genes[1] "HMGB2" "CDK1" "NUSAP1" "UBE2C" "BIRC5" "TPX2" "TOP2A"[8] "NDC80" "CKS2" "NUF2" "CKS1B" "MKI67" "TMPO" "CENPF"[15] "TACC3" "FAM64A" "SMC4" "CCNB2" "CKAP2L" "CKAP2" "AURKB"[22] "BUB1" "KIF11" "ANP32E" "TUBB4B" "GTSE1" "KIF20B" "HJURP"[29] "CDCA3" "HN1" "CDC20" "TTK" "CDC25C" "KIF2C" "RANGAP1"[36] "NCAPD2" "DLGAP5" "CDCA2" "CDCA8" "ECT2" "KIF23" "HMMR"[43] "AURKA" "PSRC1" "ANLN" "LBR" "CKAP5" "CENPE" "CTCF"[50] "NEK2" "G2E3" "GAS2L3" "CBX5" "CENPA"# 获取S期marker基因s.genes <- cc.genes$s.genes# 获取G2M期marker基因g2m.genes <- cc.genes$g2m.genes
构建Seurat对象并进行数据预处理
# Create our Seurat object and complete the initalization stepsmarrow <- CreateSeuratObject(counts = exp.mat)marrow <- NormalizeData(marrow)marrow <- FindVariableFeatures(marrow, selection.method = "vst")marrow <- ScaleData(marrow, features = rownames(marrow))
PCA降维可视化
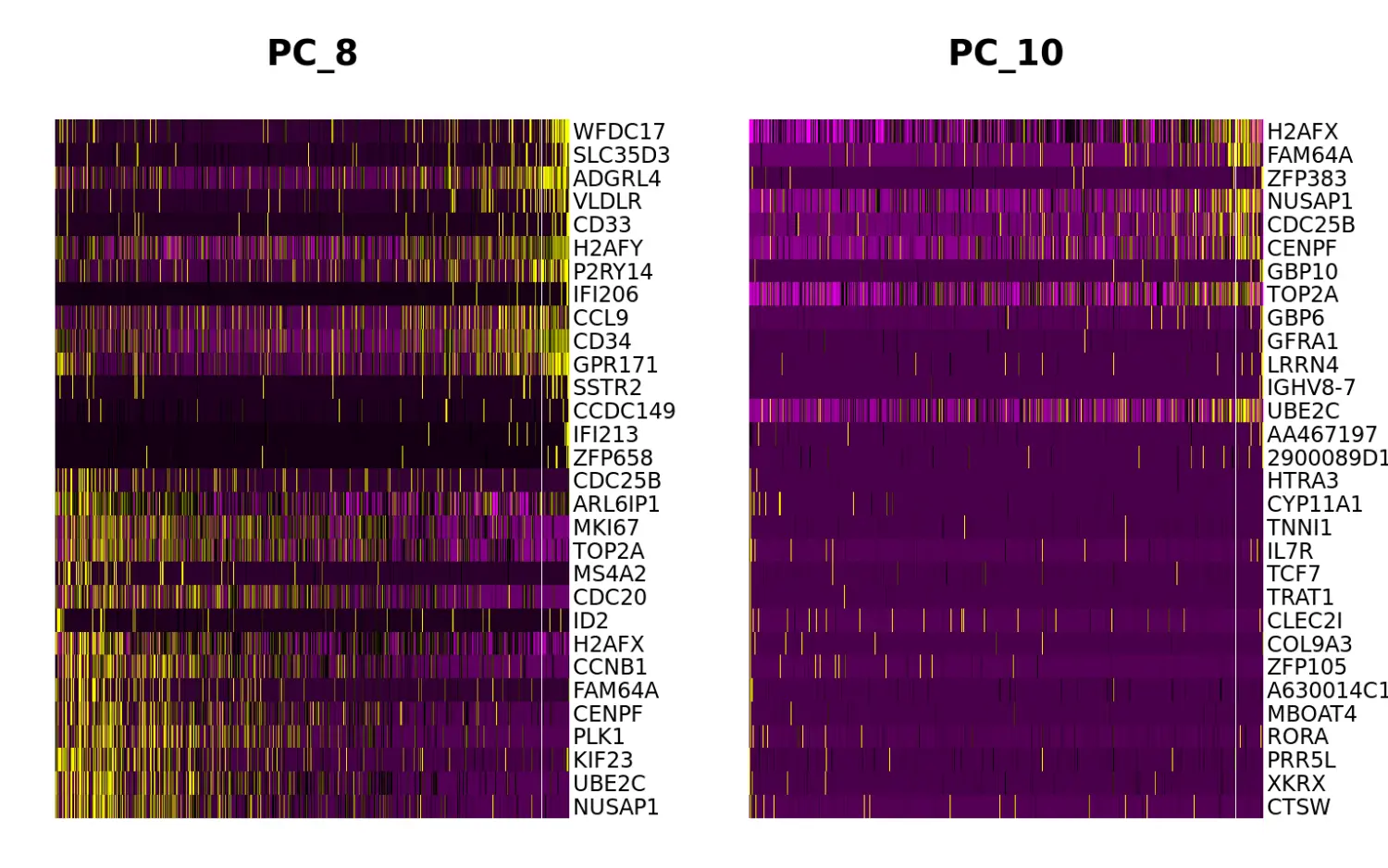
我们使用FindVariableFeatures函数筛选出的高变异基因进行PCA降维可视化,可以发现尽管大多数变异都能被解释,但是在PC8和PC10中存在一些细胞周期基因,如TOP2A和MKI67。因此,我们试图通过回归处理减少细胞周期异质性对下游分析的影响。
marrow <- RunPCA(marrow, features = VariableFeatures(marrow), ndims.print = 6:10, nfeatures.print = 10)## PC_ 6## Positive: SELL, ARL6IP1, CCL9, CD34, ADGRL4, BPIFC, NUSAP1, FAM64A, CD244, C030034L19RIK## Negative: LY6C2, AA467197, CYBB, MGST2, ITGB2, PF4, CD74, ATP1B1, GP1BB, TREM3## PC_ 7## Positive: HDC, CPA3, PGLYRP1, MS4A3, NKG7, UBE2C, CCNB1, NUSAP1, PLK1, FUT8## Negative: F13A1, LY86, CFP, IRF8, CSF1R, TIFAB, IFI209, CCR2, TNS4, MS4A6C## PC_ 8## Positive: NUSAP1, UBE2C, KIF23, PLK1, CENPF, FAM64A, CCNB1, H2AFX, ID2, CDC20## Negative: WFDC17, SLC35D3, ADGRL4, VLDLR, CD33, H2AFY, P2RY14, IFI206, CCL9, CD34## PC_ 9## Positive: IGKC, JCHAIN, LY6D, MZB1, CD74, IGLC2, FCRLA, IGKV4-50, IGHM, IGHV9-1## Negative: SLC2A6, HBA-A1, HBA-A2, IGHV8-7, FCER1G, F13A1, HBB-BS, PLD4, HBB-BT, IGFBP4## PC_ 10## Positive: CTSW, XKRX, PRR5L, RORA, MBOAT4, A630014C17RIK, ZFP105, COL9A3, CLEC2I, TRAT1## Negative: H2AFX, FAM64A, ZFP383, NUSAP1, CDC25B, CENPF, GBP10, TOP2A, GBP6, GFRA1DimHeatmap(marrow, dims = c(8, 10))
计算细胞周期评分
首先,我们基于G2/M和S期的经典marker基因的表达,计算每个细胞可能所处的细胞周期的分数。这些marker基因集应该与它们的表达水平是成反相关的关系,而那些都不表达这些marker基因的细胞可能处于G1期。
我们使用CellCycleScoring函数计算每个细胞的细胞周期得分,并将计算出的S期和G2/M期的评分保存在metadata中,以及细胞处于G2M,S或G1期的预测分类。通过设置set.ident = TRUE,则CellCycleScoring将Seurat对象中每个细胞的分组信息设置为其所处的细胞周期阶段。
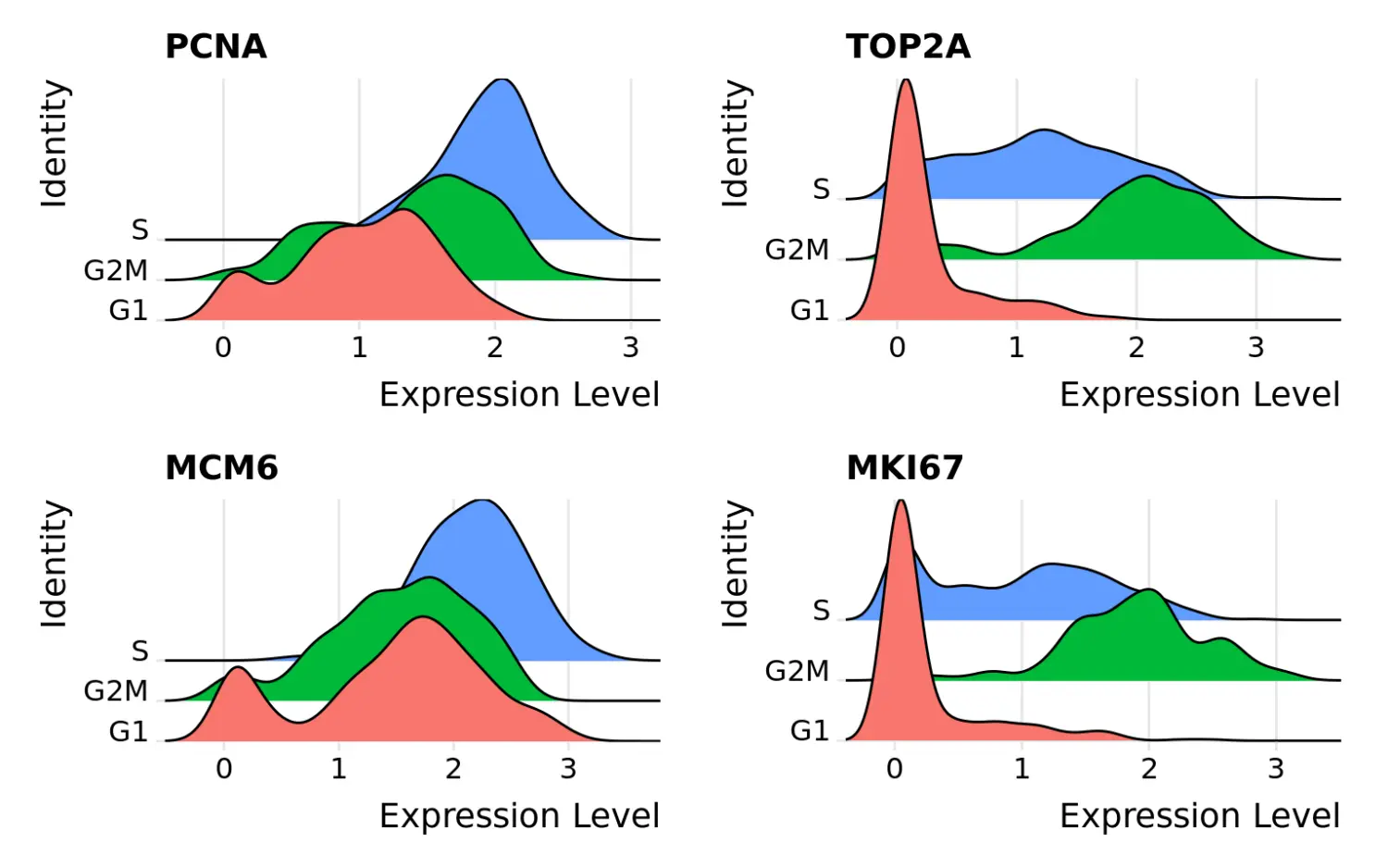
# 使用CellCycleScoring函数计算细胞周期评分marrow <- CellCycleScoring(marrow, s.features = s.genes, g2m.features = g2m.genes, set.ident = TRUE)# view cell cycle scores and phase assignmentshead(marrow[[]])orig.ident nCount_RNA nFeature_RNA S.Score G2M.Score Phase old.identProg_013 Prog 2563089 10211 -0.1424869 -0.4680395 G1 ProgProg_019 Prog 3030620 9991 -0.1691579 0.5851766 G2M ProgProg_031 Prog 1293487 10192 -0.3462704 -0.3971879 G1 ProgProg_037 Prog 1357987 9599 -0.4427021 0.6820229 G2M ProgProg_008 Prog 4079891 10540 0.5585405 0.1284359 S ProgProg_014 Prog 2569783 10788 0.0711622 0.3166073 G2M Prog# Visualize the distribution of cell cycle markers acrossRidgePlot(marrow, features = c("PCNA", "TOP2A", "MCM6", "MKI67"), ncol = 2)
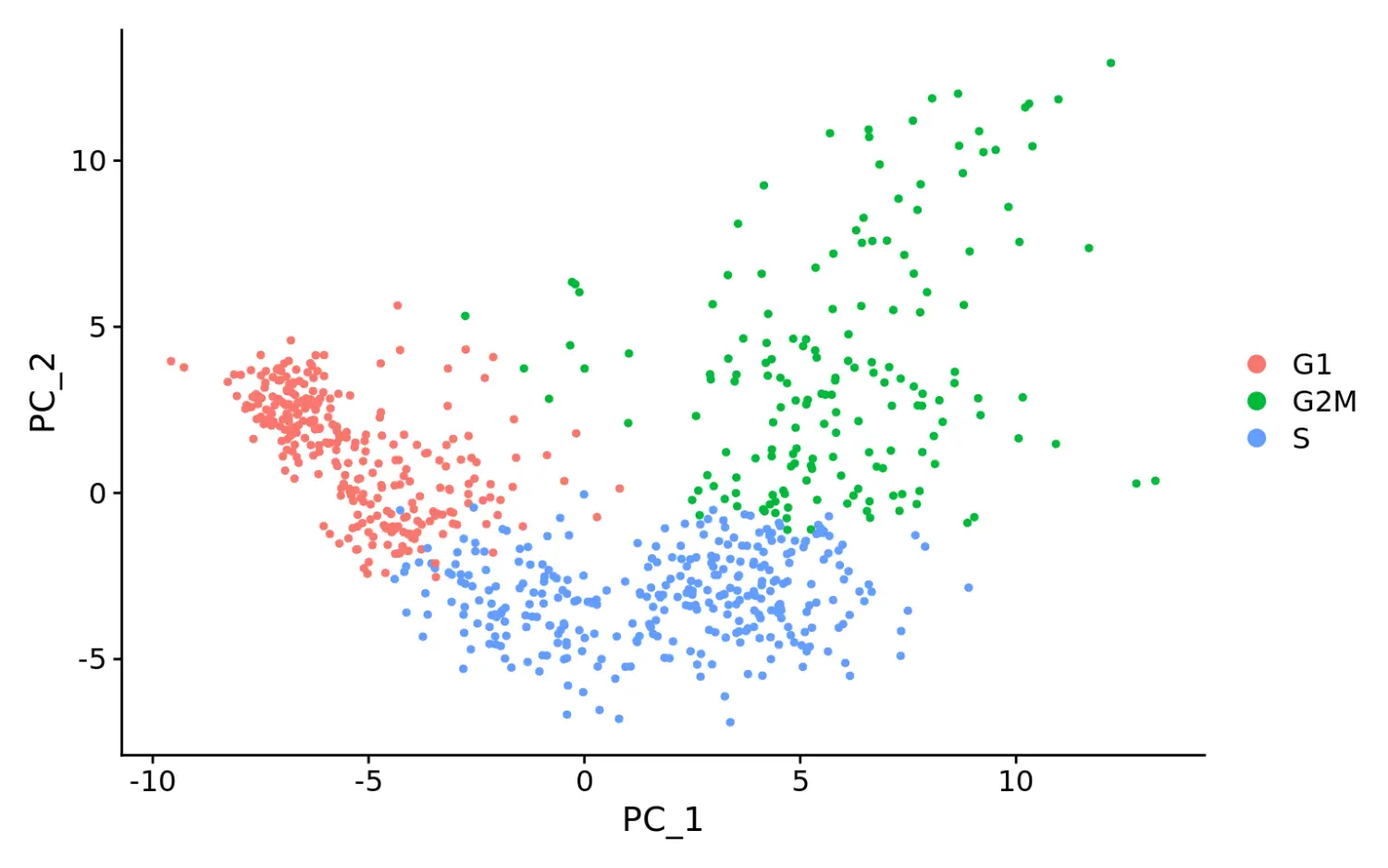
# Running a PCA on cell cycle genes reveals, unsurprisingly, that cells separate entirely by phasemarrow <- RunPCA(marrow, features = c(s.genes, g2m.genes))# 数据可视化,可以看到细胞按不同的细胞周期进行了分群DimPlot(marrow)
在数据标准化时消除细胞周期的影响
For each gene, Seurat models the relationship between gene expression and the S and G2M cell cycle scores. The scaled residuals of this model represent a ‘corrected’ expression matrix, that can be used downstream for dimensional reduction.
使用ScaleData函数进行数据标准化,并设置vars.to.regress参数指定对细胞周期评分进行回归处理,消除细胞周期异质性的影响
marrow <- ScaleData(marrow, vars.to.regress = c("S.Score", "G2M.Score"), features = rownames(marrow))# Now, a PCA on the variable genes no longer returns components associated with cell cyclemarrow <- RunPCA(marrow, features = VariableFeatures(marrow), nfeatures.print = 10)## PC_ 1## Positive: BLVRB, CAR2, KLF1, AQP1, CES2G, ERMAP, CAR1, FAM132A, RHD, SPHK1## Negative: TMSB4X, H2AFY, CORO1A, PLAC8, EMB, MPO, PRTN3, CD34, LCP1, BC035044## PC_ 2## Positive: ANGPT1, ADGRG1, MEIS1, ITGA2B, MPL, DAPP1, APOE, RAB37, GATA2, F2R## Negative: LY6C2, ELANE, HP, IGSF6, ANXA3, CTSG, CLEC12A, TIFAB, SLPI, ALAS1## PC_ 3## Positive: APOE, GATA2, NKG7, MUC13, MS4A3, RAB44, HDC, CPA3, FCGR3, TUBA8## Negative: FLT3, DNTT, LSP1, WFDC17, MYL10, GIMAP6, LAX1, GPR171, TBXA2R, SATB1## PC_ 4## Positive: CSRP3, ST8SIA6, DNTT, MPEG1, SCIN, LGALS1, CMAH, RGL1, APOE, MFSD2B## Negative: PROCR, MPL, HLF, MMRN1, SERPINA3G, ESAM, GSTM1, D630039A03RIK, MYL10, LY6A## PC_ 5## Positive: PF4, GP1BB, SDPR, F2RL2, RAB27B, SLC14A1, TREML1, PBX1, F2R, TUBA8## Negative: CPA3, LMO4, IKZF2, IFITM1, FUT8, MS4A2, SIGLECF, CSRP3, HDC, RAB44# When running a PCA on only cell cycle genes, cells no longer separate by cell-cycle phasemarrow <- RunPCA(marrow, features = c(s.genes, g2m.genes))DimPlot(marrow)
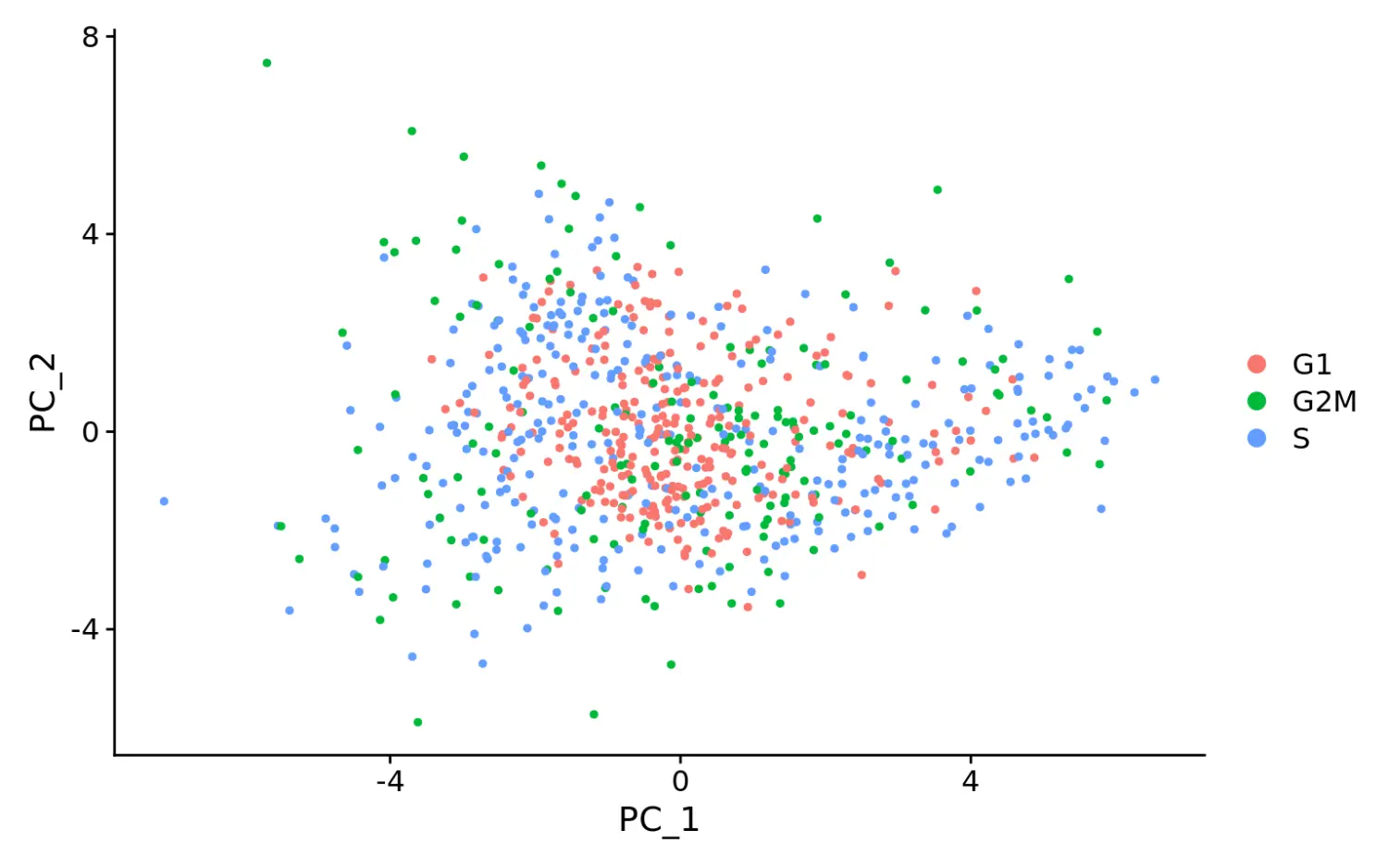
再次根据细胞周期相关基因进行PCA分析时,没有按照不同的细胞周期进行分群了,说明消除细胞周期影响的效果还是比较好的。
可选择的替代方法
在上述的分析过程中,我们消除了与细胞周期相关的所有信号。但是,在某些情况下,我们发现这会对下游的分析产生一定的负面影响,尤其是在细胞分化过程中(如鼠类造血过程)。在此过程中干细胞处于静止状态,而分化的细胞正在增殖。在这种情况下,清除所有细胞周期效应也会使干细胞和祖细胞之间的区别模糊。
作为替代方案,我们建议逐步消除G2M和S期评分之间的差异。这意味着将保持非周期细胞和周期细胞的组分差异,但是增殖细胞之间的细胞周期阶段的差异将从数据中去除。
# 计算分数差异marrow$CC.Difference <- marrow$S.Score - marrow$G2M.Score# 数据标准化消除分数差异marrow <- ScaleData(marrow, vars.to.regress = "CC.Difference", features = rownames(marrow))# cell cycle effects strongly mitigated in PCAmarrow <- RunPCA(marrow, features = VariableFeatures(marrow), nfeatures.print = 10)## PC_ 1## Positive: BLVRB, KLF1, ERMAP, FAM132A, CAR2, RHD, CES2G, SPHK1, AQP1, SLC38A5## Negative: TMSB4X, CORO1A, PLAC8, H2AFY, LAPTM5, CD34, LCP1, TMEM176B, IGFBP4, EMB## PC_ 2## Positive: APOE, GATA2, RAB37, ANGPT1, ADGRG1, MEIS1, MPL, F2R, PDZK1IP1, DAPP1## Negative: CTSG, ELANE, LY6C2, HP, CLEC12A, ANXA3, IGSF6, TIFAB, SLPI, MPO## PC_ 3## Positive: APOE, GATA2, NKG7, MUC13, ITGA2B, TUBA8, CPA3, RAB44, SLC18A2, CD9## Negative: DNTT, FLT3, WFDC17, LSP1, MYL10, LAX1, GIMAP6, IGHM, CD24A, MN1## PC_ 4## Positive: CSRP3, ST8SIA6, SCIN, LGALS1, APOE, ITGB7, MFSD2B, RGL1, DNTT, IGHV1-23## Negative: MPL, MMRN1, PROCR, HLF, SERPINA3G, ESAM, PTGS1, D630039A03RIK, NDN, PPIC## PC_ 5## Positive: GP1BB, PF4, SDPR, F2RL2, TREML1, RAB27B, SLC14A1, PBX1, PLEK, TUBA8## Negative: HDC, LMO4, CSRP3, IFITM1, FCGR3, HLF, CPA3, PROCR, PGLYRP1, IKZF2# when running a PCA on cell cycle genes, actively proliferating cells remain distinct from G1 cells however, within actively proliferating cells, G2M and S phase cells group togethermarrow <- RunPCA(marrow, features = c(s.genes, g2m.genes))DimPlot(marrow)
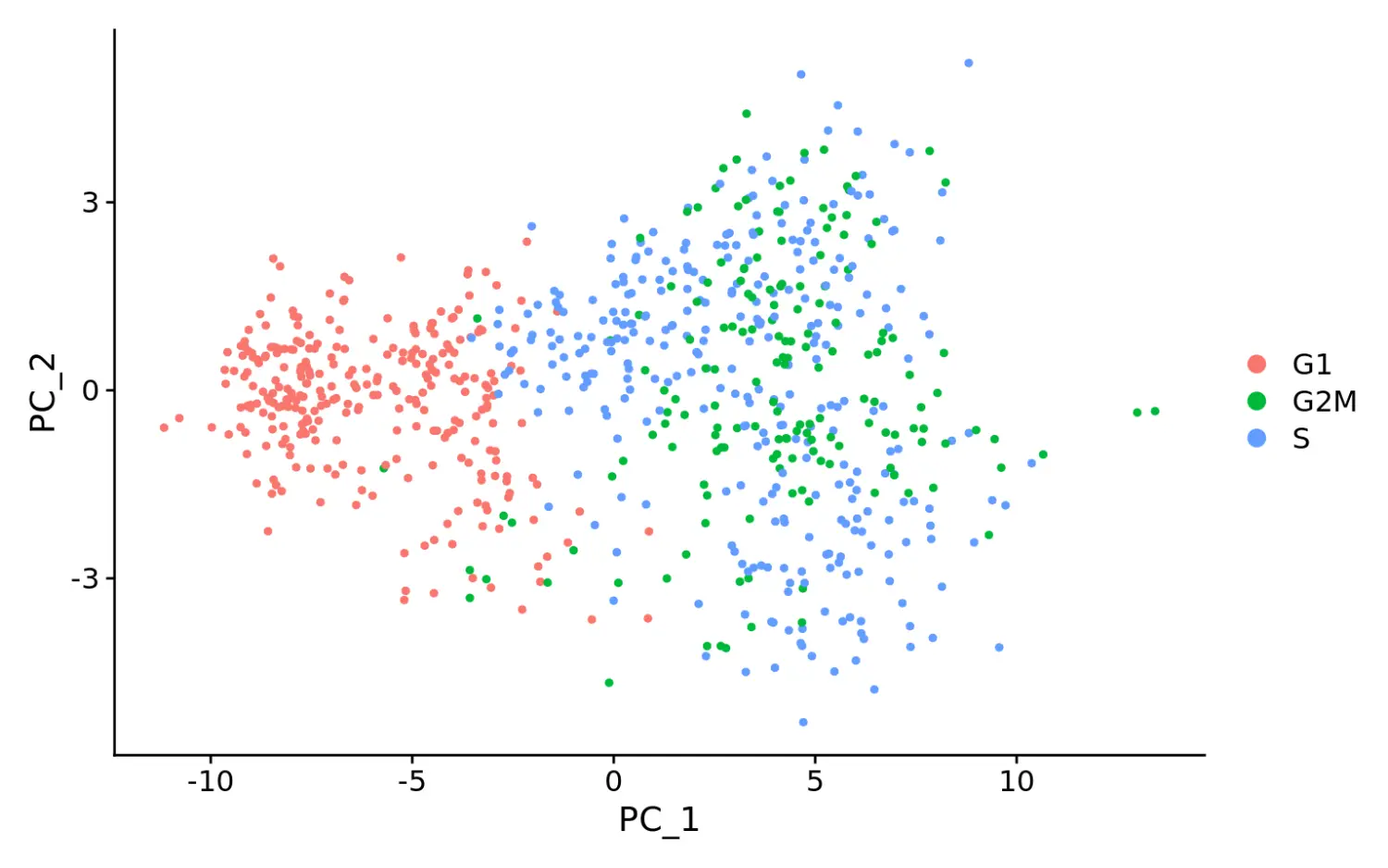
可以看到PCA降维分群后,G1期的细胞被区分开,G2/M和S期的细胞聚在一起。
参考来源:https://satijalab.org/seurat/v3.1/cell_cycle_vignette.html

