Cell Ranger ATAC简介
cellranger-atac软件是用于处理10x Genomics平台Chromium Single Cell ATAC-seq测序数据的分析流程。该软件主要包括以下四个分析流程:
- cellranger-atac mkfastq: 该子程序主要将Illumina测序仪产生的原始raw base call (BCL)测序文件转换为FASTQ文件,该命令中封装着
bcl2fastq程序。 - cellranger-atac count: 该子程序是cellranger-atac软件的主要分析流程,包括以下功能:
1)Read filtering and alignment
2)Barcode counting
3)Identification of transposase cut sites
4)Detection of accessible chromatin peaks
5)Cell calling
6)Count matrix generation for peaks and transcription factors
7)Dimensionality reduction
8)Cell clustering
9)Cluster differential accessibility - cellranger-atac aggr: 该子程序可以将多个
cellranger-atac count的分析结果进行整合处理(如,将一个实验的多个样本的分析结果进行整合),包括以下步骤:
1)Normalization of input runs to same median fragments per cell (sensitivity)
2)Detection of accessible chromatin peaks
3)Count matrix generation for peaks and transcription factors for the aggregate data
4)Dimensionality reduction
5)Cell clustering
6)Cluster differential accessibility - cellranger-atac reanalyze: 该子程序可以将
cellranger-atac count或cellranger-atac aggr的分析结果进行二次分析,可以微调一些参数进行重新分析:
1)Cell calling
2)Dimensionality reduction
3)Cell clustering
4)Cluster differential accessibility
分析流程
- One Sample, One GEM Well, One Flowcell
这是最基本的分析流程,在该分析流程中,我们只有一个生物学样本,使用一个GEM well(a set of partitioned cells from a single 10x Chromium™ Chip channel)构建单个测序文库,并使用单个flowcell进行测序。得到FASTQ测序文件后,使用cellranger-atac count子程序进行分析。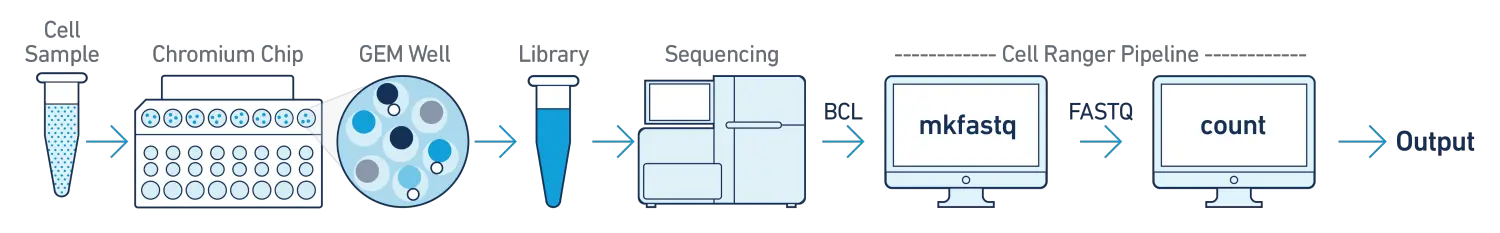
- One Sample, One GEM well, Multiple Flowcells
如果我们单个的测序文库使用多个flowcells(e.g. to increase sequencing saturation)进行测序,我们可以将不同flowcell产生的测序数据混合到一起,然后使用cellranger-atac count子程序进行分析。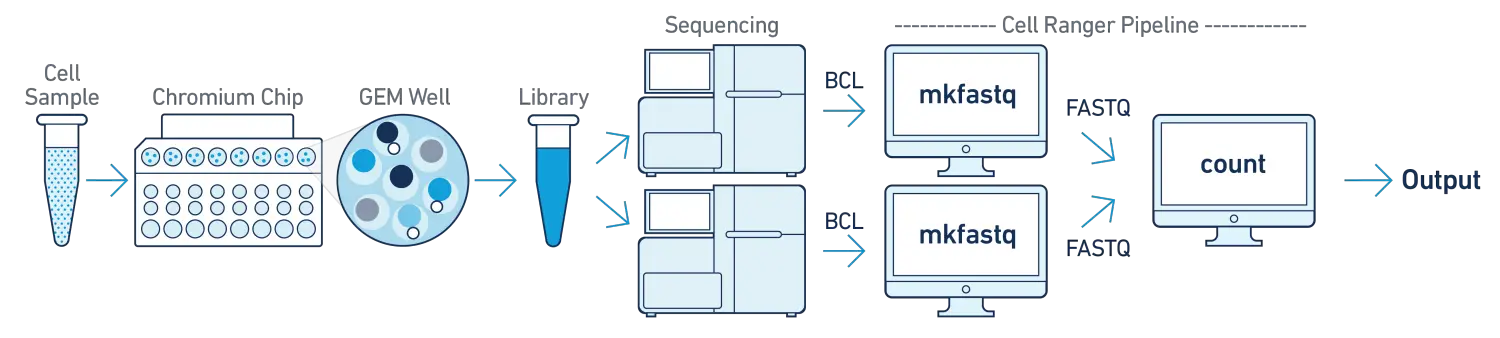
- One Sample, Multiple GEM Wells, One Flowcell
在该分析流程中,我们的单个样本使用多个不同的GEM wells进行文库构建(e.g. to conduct technical replicate experiments or to increase the number of cells in your library),然后将不同GEM wells的文库pool混合在一起,使用单个flowcell进行测序。在分析时,我们需要将混合的测序数据进行拆库demultiplex分成多个数据集,分别使用cellranger-atac count子程序进行独立分析,然后再使用cellranger-atac aggr子程序进行整合分析。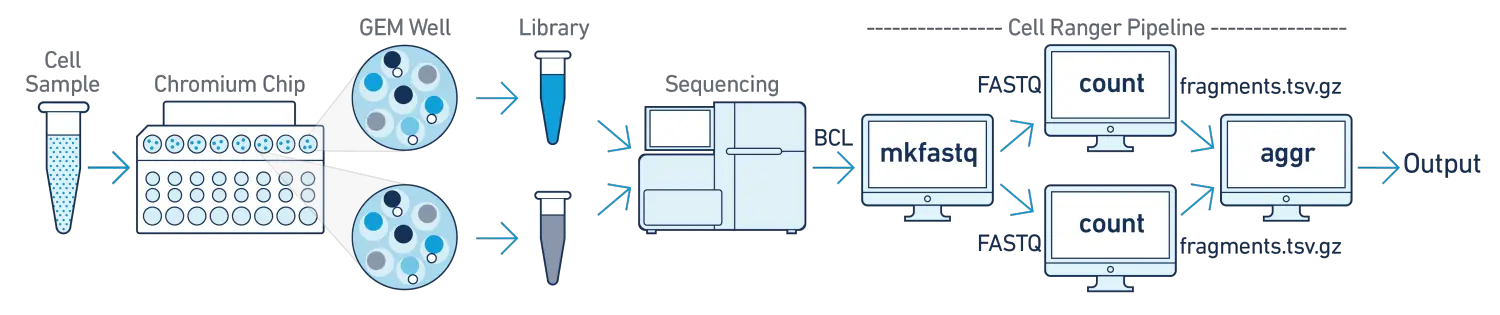
- Multiple Samples, Multiple GEM Wells, One Flowcell
在该分析流程中,我们对多个样本进行测序,每个样本分别使用不同的GEM wells进行文库构建,然后混合到一起使用单个flowcell进行测序。在分析时,我们需要将混合的测序数据进行拆库demultiplex分成多个不同样本的数据集,分别使用cellranger-atac count子程序对每个样本单独进行分析,然后可以使用cellranger-atac aggr子程序将多个样本进行整合分析。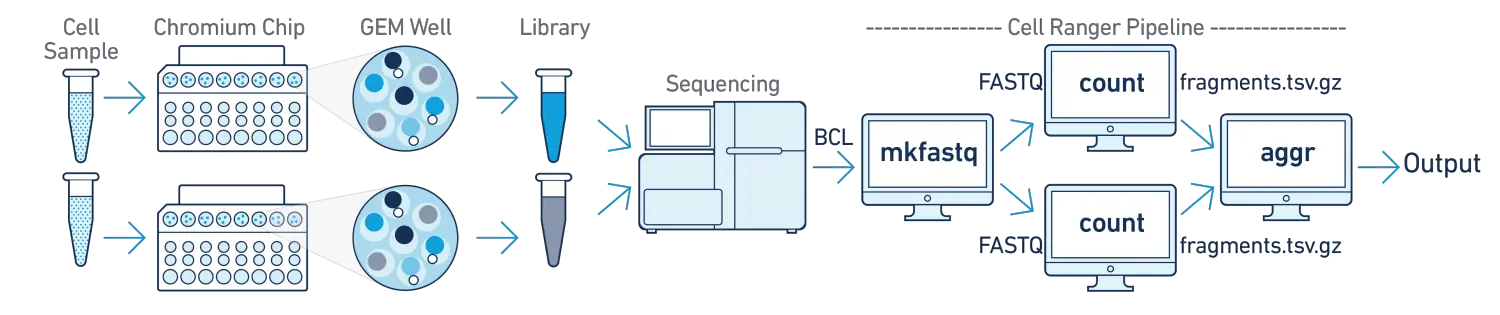
Cell Ranger ATAC 软件的下载与安装
System Requirements 系统需求
- Hardware 硬件需求
Cell Ranger ATAC pipelines run on Linux systems that meet these minimum requirements:
1)8-core Intel or AMD processor (16 cores recommended)
2)64GB RAM (128GB recommended)
3)1TB free disk space
4)64-bit CentOS/RedHat 6.0 or Ubuntu 12.04
In order to run in cluster mode, the cluster needs to meet these additional minimum requirements:
1)8-core Intel or AMD processor per node
2)6GB RAM per core
3)Shared file system (e.g. NFS)
4)SGE or LSF batch scheduling system
- Software 软件需求
In order to run cellranger-atac mkfastq, the following software needs to be installed:
1)Illumina® bcl2fastq: bcl2fastq must be version 2.17 or higher. It supports most sequencers running RTA version 1.18.54 or higher. If you are using NovaSeq™, the pipelines require version 2.20 or higher. If your sequencer is running an older version of RTA, then the pipelines require bcl2fastq 1.8.4.
- Resource Limits 系统资源需求
1)Cell Ranger ATAC runs with --jobmode=local by default, using 90% of available memory and all available cores. To restrict resource usage, please see the --localmem and --localcores flags for cellranger-atac count at the link here for more information.
2)Many Linux systems have default user limits (ulimits) for maximum open files and maximum user processes as low as 1024 or 4096. Because Cell Ranger ATAC spawns multiple processes per core, jobs that use a large number of cores can exceed these limits. 10x Genomics recommends higher limits.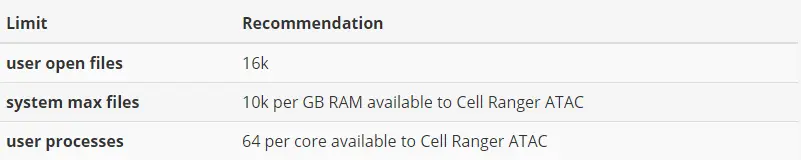
- How CPU and Memory Affect Runtime
1)运行内存的大小对软件运行时间的影响
Here is cellranger-atac count walltime as a function of available memory. In general, you can improve performance by allocating more than the minimum 64GB of memory to the pipeline. There is notable diminishing return beyond 128GB.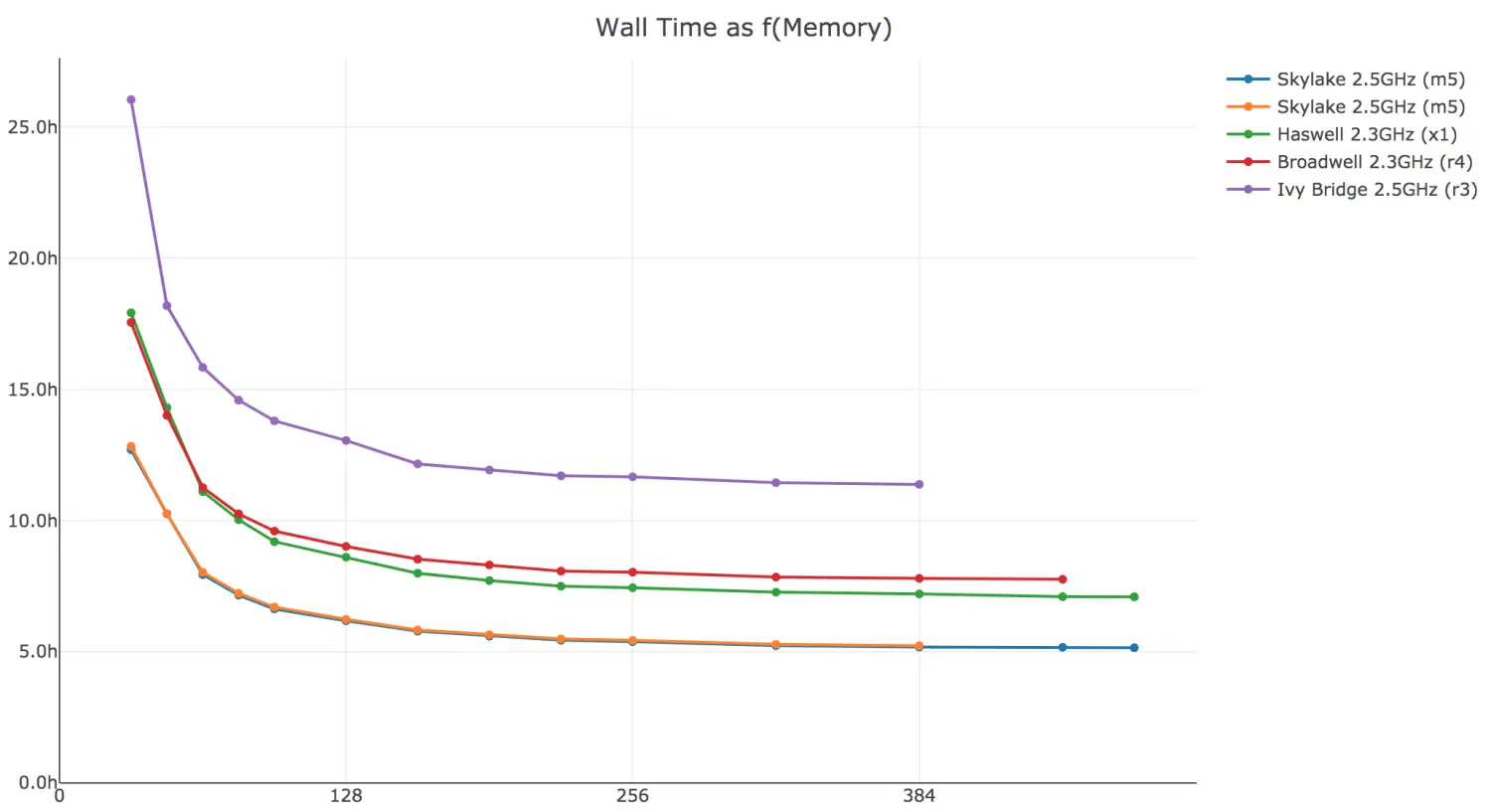
2)CPU的个数对软件运行时间的影响
Here’s cellranger-atac count walltime as a function of threads. If your system has ≫32 logical cores, you may want to run with --localcores=32 since there is diminishing return beyond 32 threads.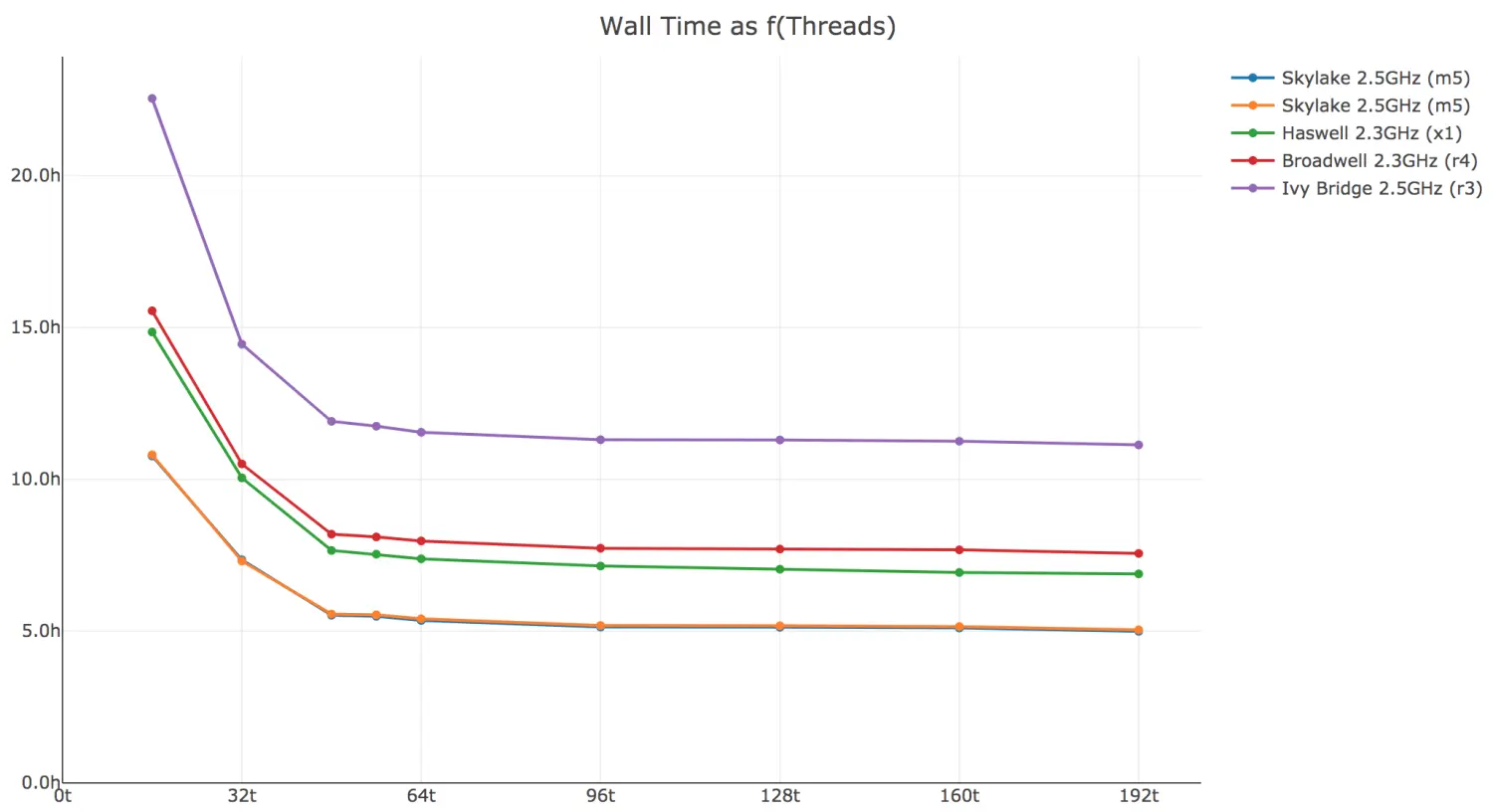
Cell Ranger ATAC Installation
Cell Ranger ATAC - 1.2.0 (November 21, 2019)(下载cellranger-atac软件)
- Self-contained, relocatable tar file. Does not require centralized installation.
- Contains binaries pre-compiled for CentOS/RedHat 6.0+ and Ubuntu 12.04+.
- Linux 64-bit – 610 MB – md5sum: 05ce0674328beb28d8f2f0b17bf1a387
# 使用curl命令下载curl -o cellranger-atac-1.2.0.tar.gz "http://cf.10xgenomics.com/releases/cell-atac/cellranger-atac-1.2.0.tar.gz?Expires=1591554210&Policy=eyJTdGF0ZW1lbnQiOlt7IlJlc291cmNlIjoiaHR0cDovL2NmLjEweGdlbm9taWNzLmNvbS9yZWxlYXNlcy9jZWxsLWF0YWMvY2VsbHJhbmdlci1hdGFjLTEuMi4wLnRhci5neiIsIkNvbmRpdGlvbiI6eyJEYXRlTGVzc1RoYW4iOnsiQVdTOkVwb2NoVGltZSI6MTU5MTU1NDIxMH19fV19&Signature=aWMB0ZS-dZgUp7RWFiN0-0lzpiervk4OULZ~igZpCJ9ojNd6c9ey3jemkayeuhEfqmQ1hYgQ4dc9wnmn383ghfVyUBW3VZljcuO3I1R5pXZ992xLWXYAFe5u6ocleEqI6LgjibRAbIInIpKwEGUTZBJrfZmddEweEZkINGkEv63e0iVWetqJj5RKuJxUP3DhGbOpBN8R7Jq0JPzp~SPDKehVCWxbBZQUp4yWGeIfCvsXOJjvAim0WAqPWsdwyW-s0m2wivOdzoeNb-XAk-ixeK45yhjJiEaqRsQl3M7rFt9EpQ2-0xrR~PO3xbTQ4T5s5kQT4VIbhhw00T9DU21~8w__&Key-Pair-Id=APKAI7S6A5RYOXBWRPDA"# or# 使用wget命令下载wget -O cellranger-atac-1.2.0.tar.gz "http://cf.10xgenomics.com/releases/cell-atac/cellranger-atac-1.2.0.tar.gz?Expires=1591554210&Policy=eyJTdGF0ZW1lbnQiOlt7IlJlc291cmNlIjoiaHR0cDovL2NmLjEweGdlbm9taWNzLmNvbS9yZWxlYXNlcy9jZWxsLWF0YWMvY2VsbHJhbmdlci1hdGFjLTEuMi4wLnRhci5neiIsIkNvbmRpdGlvbiI6eyJEYXRlTGVzc1RoYW4iOnsiQVdTOkVwb2NoVGltZSI6MTU5MTU1NDIxMH19fV19&Signature=aWMB0ZS-dZgUp7RWFiN0-0lzpiervk4OULZ~igZpCJ9ojNd6c9ey3jemkayeuhEfqmQ1hYgQ4dc9wnmn383ghfVyUBW3VZljcuO3I1R5pXZ992xLWXYAFe5u6ocleEqI6LgjibRAbIInIpKwEGUTZBJrfZmddEweEZkINGkEv63e0iVWetqJj5RKuJxUP3DhGbOpBN8R7Jq0JPzp~SPDKehVCWxbBZQUp4yWGeIfCvsXOJjvAim0WAqPWsdwyW-s0m2wivOdzoeNb-XAk-ixeK45yhjJiEaqRsQl3M7rFt9EpQ2-0xrR~PO3xbTQ4T5s5kQT4VIbhhw00T9DU21~8w__&Key-Pair-Id=APKAI7S6A5RYOXBWRPDA"
GRCh38 Reference - 1.2.0 (November 21, 2019)(下载人类hg38
参考基因组)
- Human reference (GRCh38) dataset required for Cell Ranger ATAC.
- Download – 4.9 GB – md5sum: 1b77a21f87942e069c84d2a601a41cef
curl -O http://cf.10xgenomics.com/supp/cell-atac/refdata-cellranger-atac-GRCh38-1.2.0.tar.gz# orwget http://cf.10xgenomics.com/supp/cell-atac/refdata-cellranger-atac-GRCh38-1.2.0.tar.gz
hg19 Reference - 1.2.0 (November 21, 2019)(下载人类hg19参考基因组)
- Human reference (hg19) dataset required for Cell Ranger ATAC.
- Download – 5.0 GB – md5sum: c6ff0010cc9ea628be5317594ba34ef8
curl -O http://cf.10xgenomics.com/supp/cell-atac/refdata-cellranger-atac-hg19-1.2.0.tar.gz# orwget http://cf.10xgenomics.com/supp/cell-atac/refdata-cellranger-atac-hg19-1.2.0.tar.gz
mm10 Reference - 1.2.0 (November 21, 2019)(下载小鼠mm10参考基因组)
- Mouse reference (mm10) dataset required for Cell Ranger ATAC.
- Download – 4.4 GB – md5sum: 1b71710621f0e73e37703899cae4c1bc
curl -O http://cf.10xgenomics.com/supp/cell-atac/refdata-cellranger-atac-mm10-1.2.0.tar.gz# orwget http://cf.10xgenomics.com/supp/cell-atac/refdata-cellranger-atac-mm10-1.2.0.tar.gz
软件安装
- Step 1 – Download the Cell Ranger ATAC file.
- Step 2 – Unpack the Cell Ranger ATAC file.
# 将该软件下载安装到/opt目录下cd /opt# 解压缩cellranger-atac软件包tar -xzvf cellranger-atac-1.2.0.tar.gzcd cellranger-atac-1.2.0/ls
- Step 3 – Download the reference data files.
- Step 4 – Unpack the reference data files.
# 解压缩参考基因组信息tar -xzvf refdata-cellranger-atac-GRCh38-1.2.0.tar.gzcd refdata-cellranger-atac-GRCh38-1.2.0/ls
- Step 5 – Prepend the Cell Ranger ATAC directory to your PATH. This will allow you to invoke the cellranger-atac command.
# If you unpacked both Cell Ranger ATAC and the reference data into /opt, then you would run the following command.# 将cellranger-atac软件添加到系统环境变量中export PATH=/opt/cellranger-atac-1.2.0:$PATH
You may wish to add this command to your .bashrc for convenience.
Verify Installation 检查是否安装成功
To ensure that the cellranger-atac pipeline is installed correctly, use cellranger-atac testrun. This test can take up to 60 minutes on a sixteen-core workstation. Assuming you have installed Cell Ranger ATAC into /opt, the command to run the test would look like the following:
# 将cellranger-atac软件添加到系统环境变量中export PATH=/opt/cellranger-atac-1.2.0:$PATH# 运行测试数据cellranger-atac testrun --id=tinycellranger-atac testrun 1.2.0Copyright (c) 2018 10x Genomics, Inc. All rights reserved.-------------------------------------------------------------------------------Running Cell Ranger ATAC in test mode...Martian Runtime - 3.2.4Running preflight checks (please wait)...2018-09-17 20:44:33 [runtime] (ready) ID.tiny.SC_ATAC_COUNTER_CS.SC_ATAC_COUNTER._BASIC_SC_ATAC_COUNTER._ALIGNER.SETUP_CHUNKS2018-09-17 20:44:33 [runtime] (run:local) ID.tiny.SC_ATAC_COUNTER_CS.SC_ATAC_COUNTER._BASIC_SC_ATAC_COUNTER._ALIGNER.SETUP_CHUNKS.fork0.chnk0.main...Outputs:- Per-barcode fragment counts & metrics: /opt/cellranger-atac-1.2.0/tiny/outs/singlecell.csv- Position sorted BAM file: /opt/cellranger-atac-1.2.0/tiny/outs/possorted_bam.bam- Position sorted BAM index: /opt/cellranger-atac-1.2.0/tiny/outs/possorted_bam.bam.bai- Summary of all data metrics: /opt/cellranger-atac-1.2.0/tiny/outs/summary.json- HTML file summarizing data & analysis: /opt/cellranger-atac-1.2.0/tiny/outs/web_summary.html- Bed file of all called peak locations: /opt/cellranger-atac-1.2.0/tiny/outs/peaks.bed- Raw peak barcode matrix in hdf5 format: /opt/cellranger-atac-1.2.0/tiny/outs/raw_peak_bc_matrix.h5- Raw peak barcode matrix in mex format: /opt/cellranger-atac-1.2.0/tiny/outs/raw_peak_bc_matrix- Directory of analysis files: /opt/cellranger-atac-1.2.0/tiny/outs/analysis- Filtered peak barcode matrix in hdf5 format: /opt/cellranger-atac-1.2.0/tiny/outs/filtered_peak_bc_matrix.h5- Filtered peak barcode matrix in mex format: /opt/cellranger-atac-1.2.0/tiny/outs/filtered_peak_bc_matrix- Barcoded and aligned fragment file: /opt/cellranger-atac-1.2.0/tiny/outs/fragments.tsv.gz- Fragment file index: /opt/cellranger-atac-1.2.0/tiny/outs/fragments.tsv.gz.tbi- Filtered tf barcode matrix in hdf5 format: /opt/cellranger-atac-1.2.0/tiny/outs/filtered_tf_bc_matrix.h5- Filtered tf barcode matrix in mex format: /opt/cellranger-atac-1.2.0/tiny/outs/filtered_tf_bc_matrix- Loupe Cell Browser input file: /opt/cellranger-atac-1.2.0/tiny/outs/cloupe.cloupe- csv summarizing important metrics and values: /opt/cellranger-atac-1.2.0/tiny/outs/summary.csv- Annotation of peaks with genes: /opt/cellranger-atac-1.2.0/tiny/outs/peak_annotation.tsvPipestance completed successfully!Saving diagnostics to tiny/tiny.mri.tgz# 查看测试数据运行的结果cd tiny/outsls

Diganostics will be saved whether the test succeeds or fails. This tiny.mri.tgz file contains diagnostic information 10x Genomics can use to help resolve any problems. If the pipeline fails and you need troubleshooting assistance, you can send this file directly to us from the command line.
cellranger-atac upload your@email.edu tiny/tiny.mri.tgz
参考来源:https://support.10xgenomics.com/single-cell-atac/software/pipelines/latest/installation

