效果如下:
记录几个功能,首先是放大区域:
ggplot(data_matrix, aes(gene, exon)) +geom_point() + geom_smooth(method = "lm") +geom_point(data=subset( data_matrix,gene < 2.5 & exon > (0.936*gene + 0.4094)),color = "red") +facet_zoom(x = group == "marked")
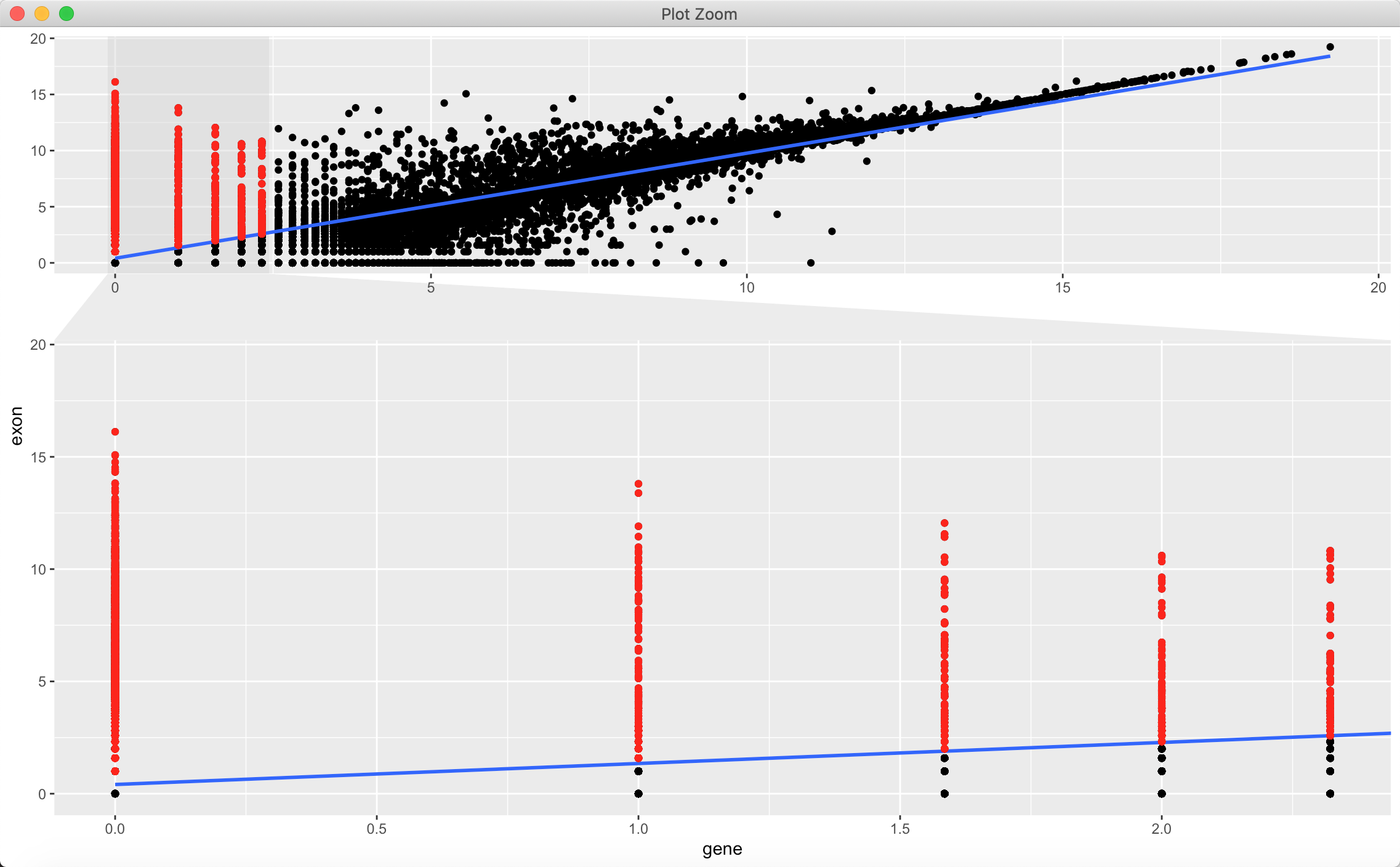
x, y 控制放大是在横坐标还是纵坐标。
此外,还可以绘制选择图片的区域轮廓:
参见,https://www.cnblogs.com/Mao1518202/p/11961495.html
比如:
ggplot(data_matrix, aes(gene, exon)) +geom_point() + geom_smooth(method = "lm") +geom_point(data=subset( data_matrix,gene < 2.5 & exon > (0.936*gene + 0.4094)),color = "red") +geom_mark_ellipse(data=subset( data_matrix,gene < 2.5 & exon > (0.936*gene + 0.4094)),aes(label = group), color = "red")
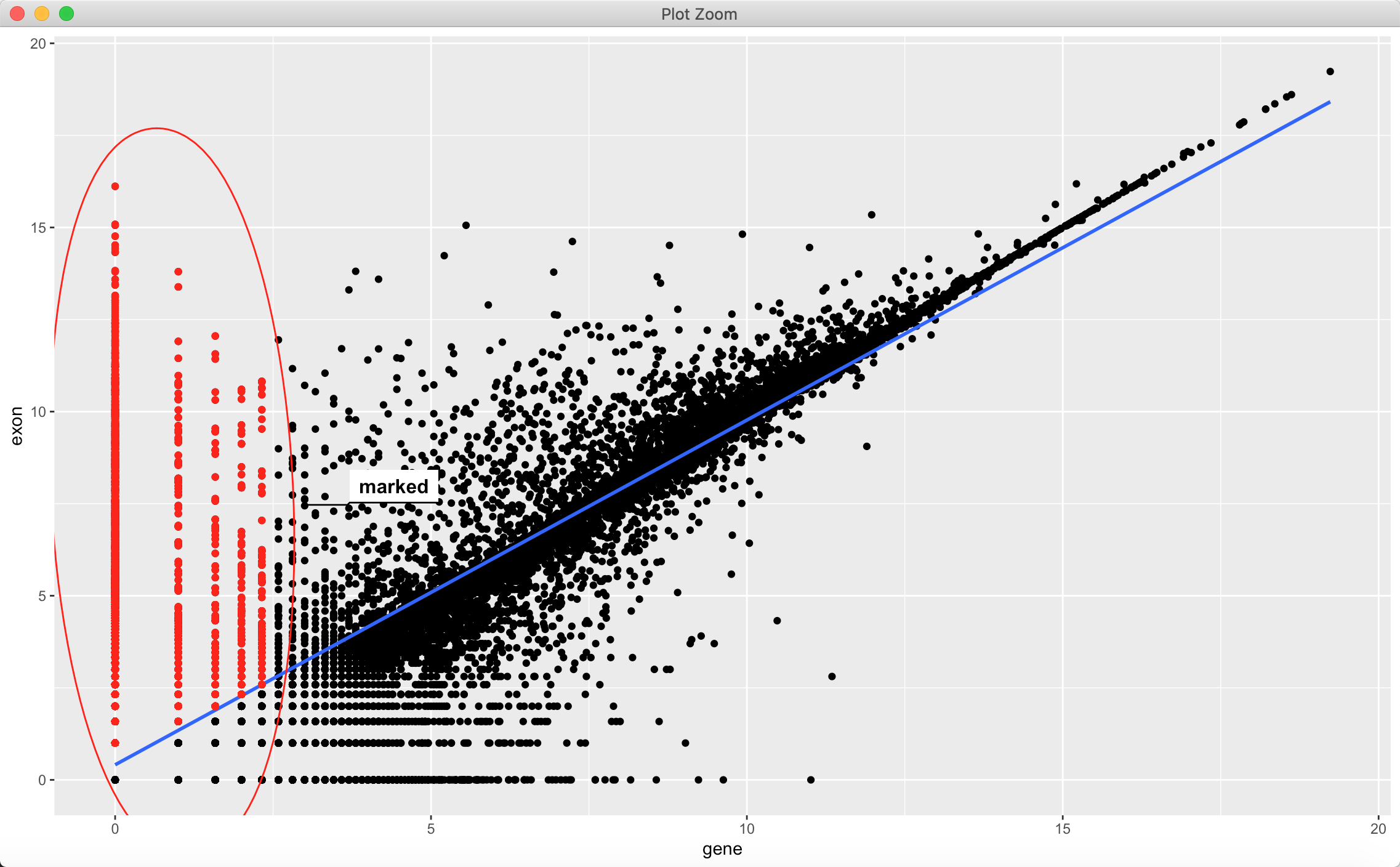
但如果不是这样的连续性数据呢?
比如分类的一个个散点(气泡图):
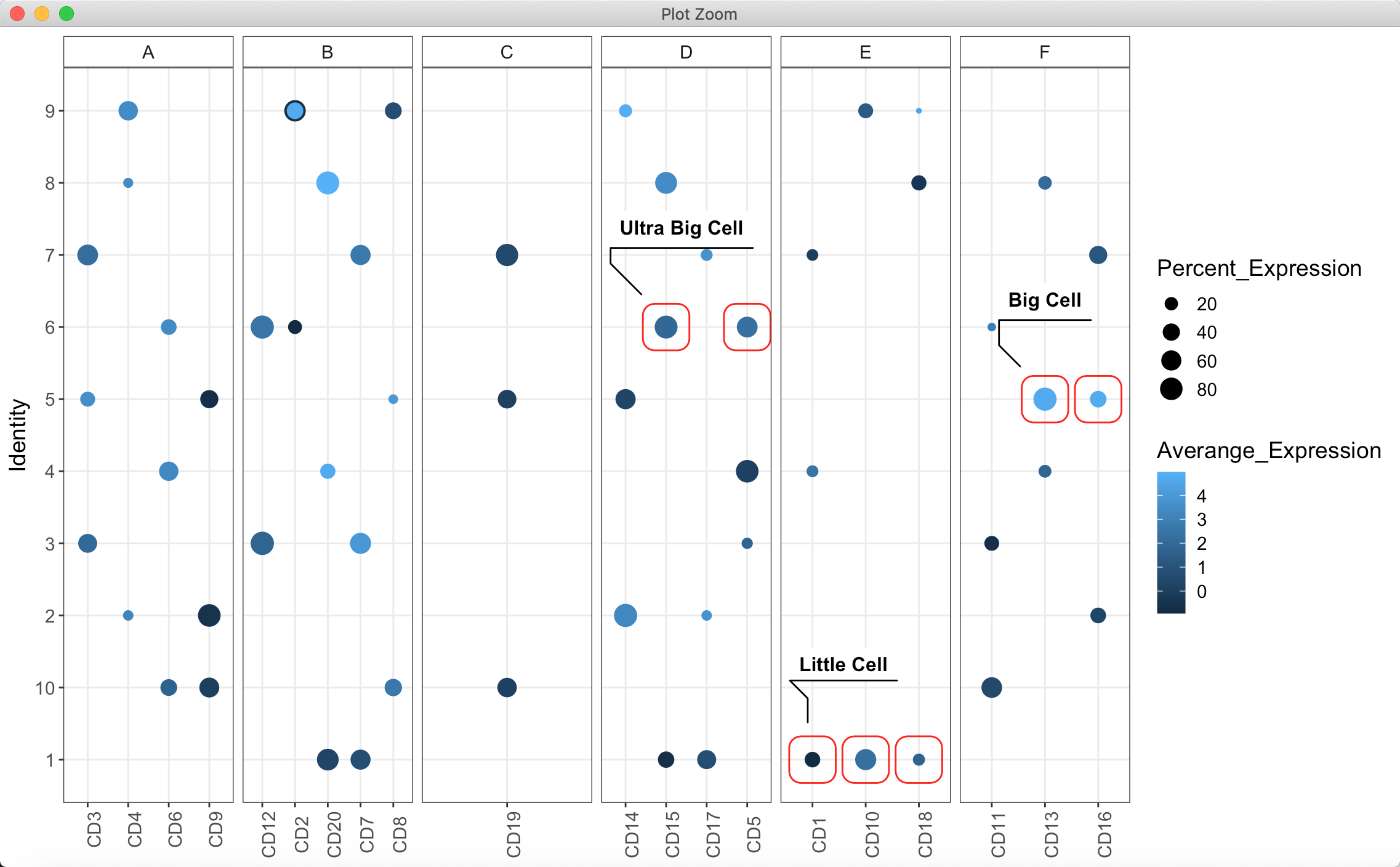
发现并不能很好的识别出相邻的数据(每个点都是单独的一个圈圈),只能实现上面的效果啦~
my_test <- data.frame(Features = rep(paste0("CD", 1:20), 3),Identity = as.character(sample(10, 60 ,replace = T)),Averange_Expression = runif(60, -1, 5),Percent_Expression = runif(60, 0, 90))tmp <- data.frame(Features = paste0("CD", 1:20),Big_Features = as.character(sample(LETTERS[1:6], 20 ,replace = T)))my_test <- merge(my_test, tmp, by = "Features")my_test$Labels <- case_when(my_test$Features %in% c("CD13", "CD16") & my_test$Identity == "5" ~ "Big Cell",my_test$Features %in% c("CD15", "CD5") & my_test$Identity == "6" ~ "Ultra Big Cell",my_test$Features %in% c("CD1", "CD10", "CD18") & my_test$Identity == "1" ~ "Little Cell",TRUE ~ "no-marked")my_test[!my_test$Labels %in% "no-marked" & duplicated(my_test$Labels), ]$Labels <- NAggplot(my_test) + geom_point(aes(Features, Identity,color = Averange_Expression,size = Percent_Expression)) +theme_bw() + facet_grid(.~Big_Features, scales = "free") +theme(axis.title.x = element_blank(),axis.text.x = element_text(angle = 90),text = element_text(size = 14),strip.background = element_rect(fill = "transparent")) +geom_mark_rect(data = my_test[(!my_test$Labels %in% "no-marked") & (!is.na(my_test$Labels)), ],aes(Features, Identity, label = Labels),color = "red") +geom_mark_rect(data = my_test[(!my_test$Labels %in% "no-marked") & (is.na(my_test$Labels)), ],aes(Features, Identity),color = "red")
有大神的话,可以帮帮忙~

