文章标题:Tet inactivation disrupts YY1 binding and long-range chromatin interactions during embryonic heart development
获取地址:https://www.nature.com/articles/s41467-019-12325-z#Sec11
Nature Communications volume 10, Article number: 4297 (2019)
Abstract
Tet-mediated DNA demethylation plays an important role in shaping the epigenetic landscape and chromatin accessibility to control gene expression. While several studies demonstrated pivotal roles of Tet in regulating embryonic development, little is known about their functions in heart development. Here we analyze DNA methylation and hydroxymethylation dynamics during early cardiac development in both human and mice. We find that cardiac-specific deletion of Tet2 and Tet3 in mice (Tet2/3-DKO) leads to ventricular non-compaction cardiomyopathy (NCC) with embryonic lethality. Single-cell RNA-seq analyses reveal a reduction in cardiomyocyte numbers and transcriptional reprogramming in cardiac tissues upon Tet2/3 depletion. Impaired DNA demethylation and reduced chromatin accessibility in Tet2/3-DKO mice further compromised Ying-yang1 (YY1) binding to its genomic targets, and perturbed high-order chromatin organization at key genes involved in heart development. Our studies provide evidence of the physiological role of Tet in regulating DNA methylation dynamics and chromatin organization during early heart development.
【TET介导的DNA去甲基化在塑造表观遗传景观和染色质可用性以控制基因表达中起着重要作用。 虽然几项研究表明Tet在调节胚胎发育方面的关键作用,但对其在心脏发育中的功能知之甚少。 在这里,我们在人和小鼠的早期心脏发育过程中分析DNA甲基化和羟甲基化动力学。 我们发现小鼠(TET2 / 3-DKO)中TET2和TET3的心脏特异性缺失导致心室不压实心肌病(NCC),具有胚胎致死性。 单细胞RNA-SEQ分析揭示了在TET2 / 3耗尽时心肌组织数和心脏组织转录重编程的降低。 DNA去甲基化和TET2 / 3-DKO小鼠中的DNA去甲基化和染色质取证性降低进一步受到ying-Yang1(YY1)与其基因组靶标结合,并在涉及心脏发育的关键基因中扰动了高阶染色质组织。 我们的研究提供了TET在早期心脏发育期间调节DNA甲基化动力学和染色质组织的生理作用的证据。】
Methods
WGBS library construction and data analysis
Purified genomic DNA (with 5% of unmethylated lambda DNA spike-in, Promega) was sheared to till reaching a fragment size of 200–500 bp using Bioruptor UCD300 (Diagenode) according to manufacturer’s instructions. Sheared DNA was ligated with methylated adaptors (NEBNext® Multiplex Oligos for Illumina®, NEB) by using a NEBNext® Ultra™ II DNA Library Prep Kit (NEB). Methylated adaptor-ligated DNA fragment was used for bisulfited conversion reaction with EZ DNA Methylation-Lightning Kit (Zymo Research), then bisulfite converted DNA was amplified using KAPA HiFi HotStart Uracil + ReadyMix PCR Kit (Kapa Biosystems) with 8 cycles of PCR. Amplified DNA was purified by AMPureXP beads and examined by Agilent High Sensitivity DNA kit (Agilent Technologies) for quality check. Library concentration was determined by a Qubit 4 fluorometer (Thermo Fisher Scientific). Prepared libraries were sequenced using an Illumina NextSeq 500 instrument (150-cycle, paired-end).
Raw fastq files for WGBS (from E10.5-P0 stage) were downloaded from ENCODE. Raw fastq files were mapped to the hg19/mm10 genome assembly using bsmap-2.89 software with “-v 6 -n 1 -q 3 -r 1” parameters. The bisulfite conversion ratios were estimated using unmethylated lambda DNA. Mcall modual in MOABS was used to call the mCG/CG ratios for each CpG site. Mcomp modual was used to call DMRs with parameter “–minNominalDif = 0.2–minDmcsInDmr 3–maxDistConsDmcs 500”. The CpGs with coverage > = 5 was used for downstream analysis. The function prediction of DMRs was used for GREAT analysis. UCSC genome browser tracks were generated by using the Mmint ucsc.py function.
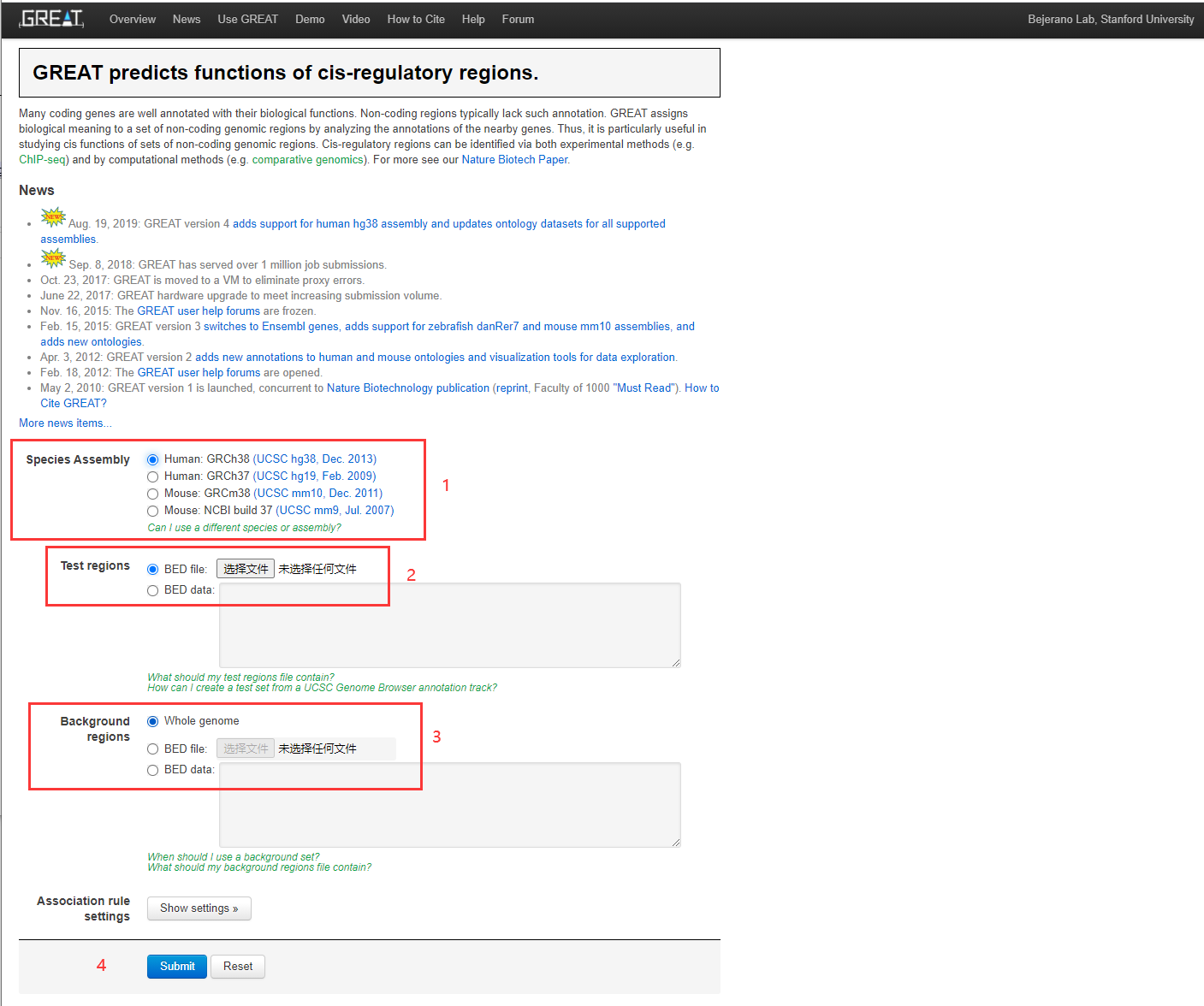
GREAT预测富集非编码区域
文章中使用到了“GREAT”这个在线分析网站来分析差异甲基化区域,感觉比较新颖,因为没用过,现在记录一下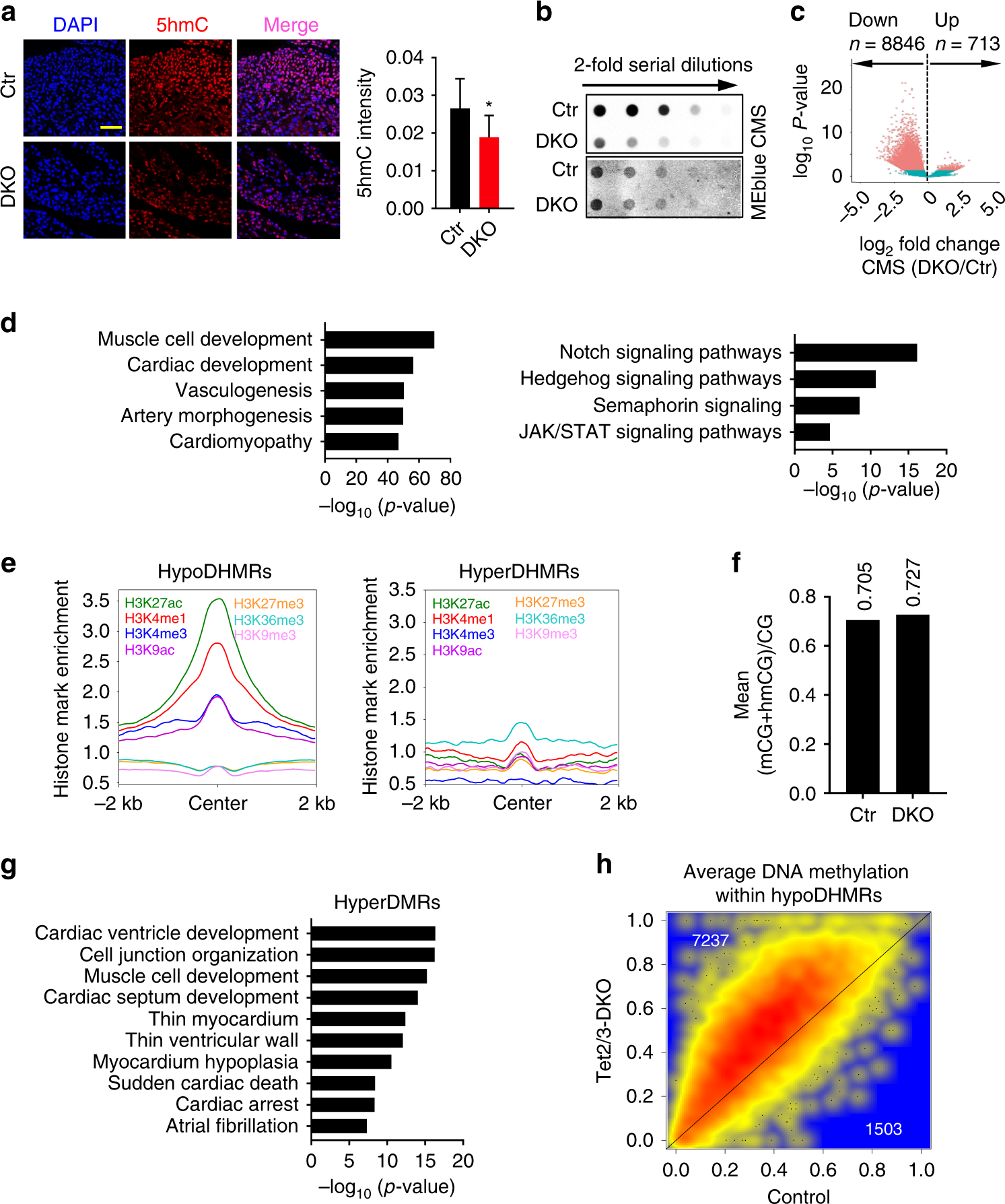
作者使用
Tet2/3 deletion in embryonic heart resulted impaired 5hmC but not 5mC.
a (Left) Representative IF staining images for control and Tet2/3-DKO murine heart tissues collected at E12.5. Blue, DAPI; Red, 5hmC. (Right) Quantification of 5hmC levels in control and Tet2/3-DKO heart tissues collected at E12.5. Data were shown as mean ± S.D; n = 3 independent experiments (a total of 1038 and 1250 cells were analyzed for control and Tet2/3-DKO, respectively). **p < 0.01 compared to control (two-tailed Student’s t-test were used). Scale bar: 50 µm.
b Measurement of the global 5hmC levels in control and Tet2/3-DKO heart tissues collected at E12.5 by using the dot-blot assay. Methylene blue (MEblue; bottom) staining was used to visualize the total DNA input.
c Volcano plot illustrating the differentially enriched 5hmC regions (DHMRs) in E12.5 heart tissues between the control and Tet2/3-DKO groups (p value < = 0.05). The mean size of hypoDHMRs was 593 bp, covering 0.18% of the genome. 【在对照和TET2 / 3-DKO基团之间的E12.5心脏组织中差异富集的5hmC区(DhMRs)的火山图(P值<= 0.05)。 HypodhMR的平均尺寸为593bp,占该基因组的0.18%。】
d Representative GREAT analysis on hypoDHMRs illustrated in Fig. 4c. Corrected binomial raw p-value were calculated. 【hypoDHMRs的典型GREAT分析如图4c所示。计算校正后的二项原始p值。】
e Normalized enrichment of the indicated histone modifications within identified hypoDHMRs (left) and hyperDHMRs (right). 【在确定的hypoDHMRs(左)和hyperDHMRs(右)中指示组蛋白修饰的标准化富集】
f Average DNA methylation levels in control and Tet2/3-DKO murine heart tissues collected at E12.5. 【在E12.5收集的对照和TET2 / 3-DKO鼠心脏组织的平均DNA甲基化水平。】
g GREAT analyses on identified top 5000 most significant changed hyperDMRs in Tet2/3-DKO heart tissues compared with control. Corrected binomial raw p-value were calculated. 【与对照相比,对TET2 / 3-DKO心脏组织中鉴定的前5000个最显着改变的Hyperdmrs的GREAT分析。 计算校正的二项式原始p值。】
h Scatterplot of the average DNA methylation levels within hypoDHMRs in E12.5 control and Tet2/3-DKO heart tissues.【E12.5对照和TET2 / 3-DKO心脏组织中hypoDHMRs【什么是DhMRs】内平均DNA甲基化水平的散点图】
什么是GREAT,这个工具能做什么?
Many coding genes are well annotated with their biological functions. Non-coding regions typically lack such annotation. GREAT assigns biological meaning to a set of non-coding genomic regions by analyzing the annotations of the nearby genes. Thus, it is particularly useful in studying cis functions of sets of non-coding genomic regions. Cis-regulatory regions can be identified via both experimental methods (e.g. ChIP-seq) and by computational methods (e.g. comparative genomics). For more see our Nature Biotech Paper(http://bejerano.stanford.edu/papers/GREAT.pdf).
地址:http://great.stanford.edu/public/html/
标签:GREAT predicts functions of cis-regulatory regions.