Arrowhead(https://github.com/aidenlab/juicer/wiki/Arrowhead)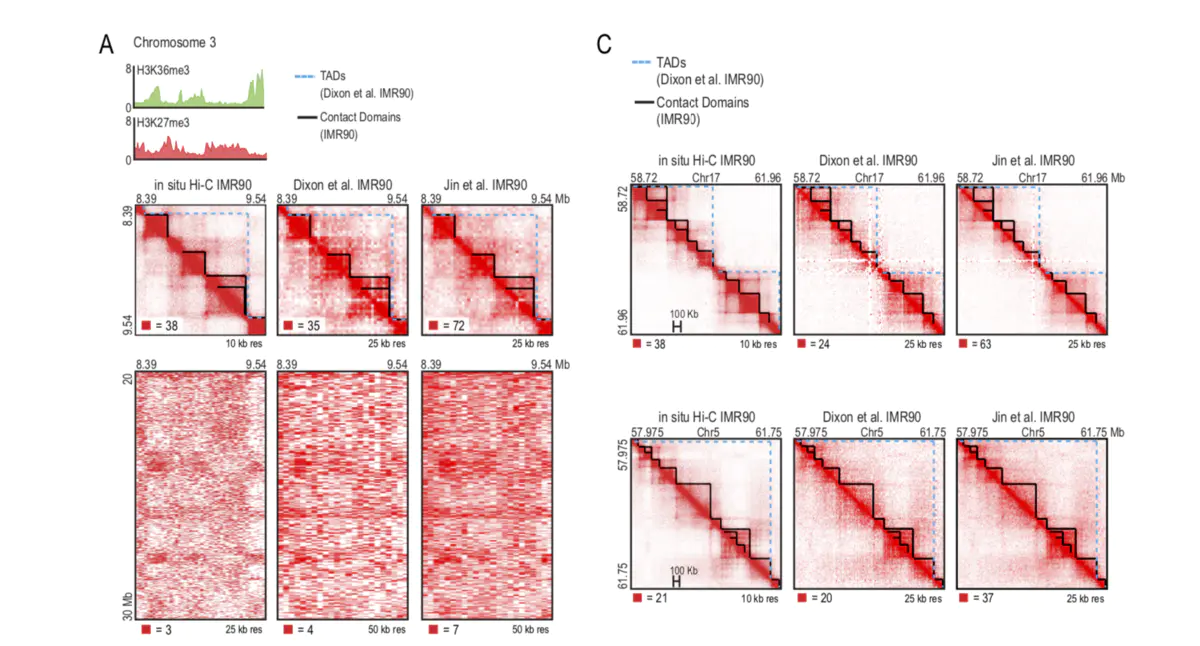
在以往的calling TAD 的软件比较中,往往把arrowhead和其他软件放入其中一同比较。但阅读原文发现arrowhead 并非用来鉴定TAD,而是一些concat domain。这些contact domain 有些与CTCF的loop相对应,而其他的更像是sub TAD(如下图所示)。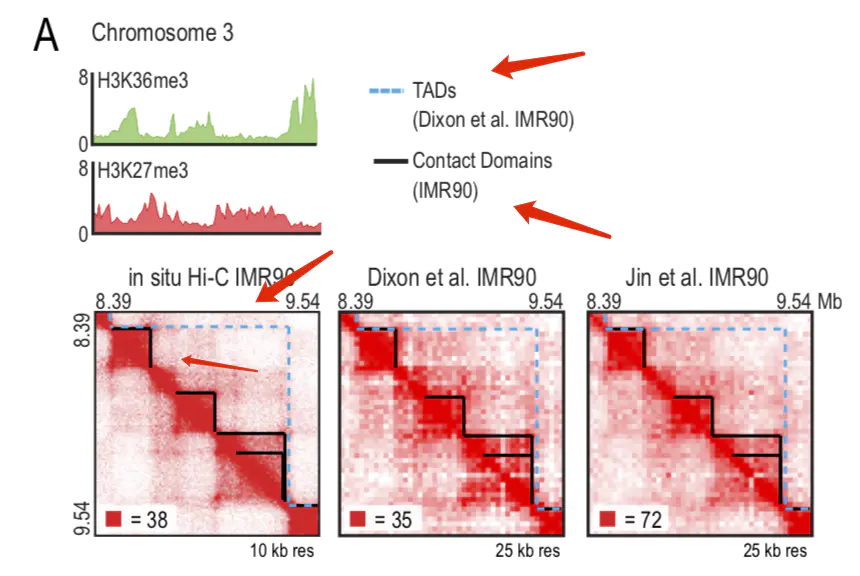
让我们再来看看原文的描述:
In our new, higher resolution maps (200- to 1,000-fold more contacts), we observe many small squares of enhanced contact frequency that tile the diagonal of each contact matrix (Figure 2A). We used the Arrowhead algorithm (see Experimental Procedures) to annotate these contact domains genome-wide. The observed domains ranged in size from 40 kb to 3 Mb (median size 185 kb). As with megadomains, there is an abrupt drop in contact frequency (33%) for pairs of loci on opposite sides of the domain boundary (Figure S2G). Contact domains are often preserved across cell types (Figures S3A and S3B).
看完了arrowhead的描述,让我们看看脚本:
juicer_tools arrowhead sam1_chr10.hic ./contact_domains_list/
arrowhead 脚本非常简单只需要输入.hic 文件和输出路径即可。
arrowhead 结果文件:
chr1 x1 x2 chr2 y1 y2 name score strand1 strand2 color score uVarScore lVarScore upSign loSign10 52700000 53985000 10 52700000 53985000 . . . . 255,255,0 0.41747454097878034 0.41970000160383647 0.409477935545163 0.43657232137491736 0.4802595997836668410 9365000 11530000 10 9365000 11530000 . . . . 255,255,0 0.36420329490041414 0.41362580689849504 0.3894973896850802 0.401325150247403840.464057423177387510 15455000 16850000 10 15455000 16850000 . . . . 255,255,0 0.5489743298631723 0.40563766266026124 0.35380396518498225 0.476734693877551 0.588214285714285710 125745000 126090000 10 125745000 126090000 . . . . 255,255,0 0.6609615269302085 0.36916154242309135 0.3879633742571127 0.5828571428571429 0.554285714285714310 100215000 101135000 10 100215000 101135000 . . . . 255,255,0 0.755689961290954 0.31256138469800887 0.3220336759786886 0.5375175315568023 0.583917718560074810 6450000 7525000 10 6450000 7525000 . . . . 255,255,0 0.8279503126182577 0.3265788783197742 0.30777127122093495 0.5869341563786008 0.608110425240054910 15920000 16830000 10 15920000 16830000 . . . . 255,255,0 0.9782706809375372 0.30248721264752976 0.24512848132657034 0.5738174868609651 0.751433349259436210 96370000 96870000 10 96370000 96870000 . . . . 255,255,0 0.900385516954811 0.24702025315833226 0.28021307721486366 0.7490196078431373 0.454117647058823510 50785000 51125000 10 50785000 51125000 . . . . 255,255,0 0.9495222201309195 0.3058902380876848 0.19212681234041373 0.6067226890756302 0.7605042016806722
每列代表如下:
- chromosome = the chromosome that the domain is located on x1,x2/y1,y2 = the interval spanned by the domain (contact domains manifest as squares on the diagonal of a Hi-C matrix and as such: x1=y1, x2=y2)
- color = the color that the feature will be rendered as if loaded in Juicebox
- corner_score = the corner score, a score indicating the likelihood that a pixel is at the corner of a contact domain. Higher values indicate a greater likelihood of being at the corner of a domain
- Uvar = the variance of the upper triangle
- Lvar = the variance of the lower triangle
- Usign = -1*(sum of the sign of the entries in the upper triangle)
- Lsign = sum of the sign of the entries in the lower triangle
作者:XuningFan
链接:https://www.jianshu.com/p/800d671e70fa
来源:简书
著作权归作者所有。商业转载请联系作者获得授权,非商业转载请注明出处。

