做生信的童鞋想要学习 Docker,或者使用 Docker+Pipeline 封装自己的一套数据分析流程,相信一定不能错过胡博强老师在2017年写这篇《[Docker]使用阿里云 + Docker 分析高通量测序数据——RNA-Seq 与 ChIP-Seq. - Boqiang Hu》教程,这个教程同时也以推文的方式发布在了 2017-03-21 生信技能树公众号上,感兴趣的同学可以自己去翻一下。
根据教程+tangEpiNGSInstall 仓库提供的原始测试数据,本人这两天测试跑了一下,发现了一点点小问题。
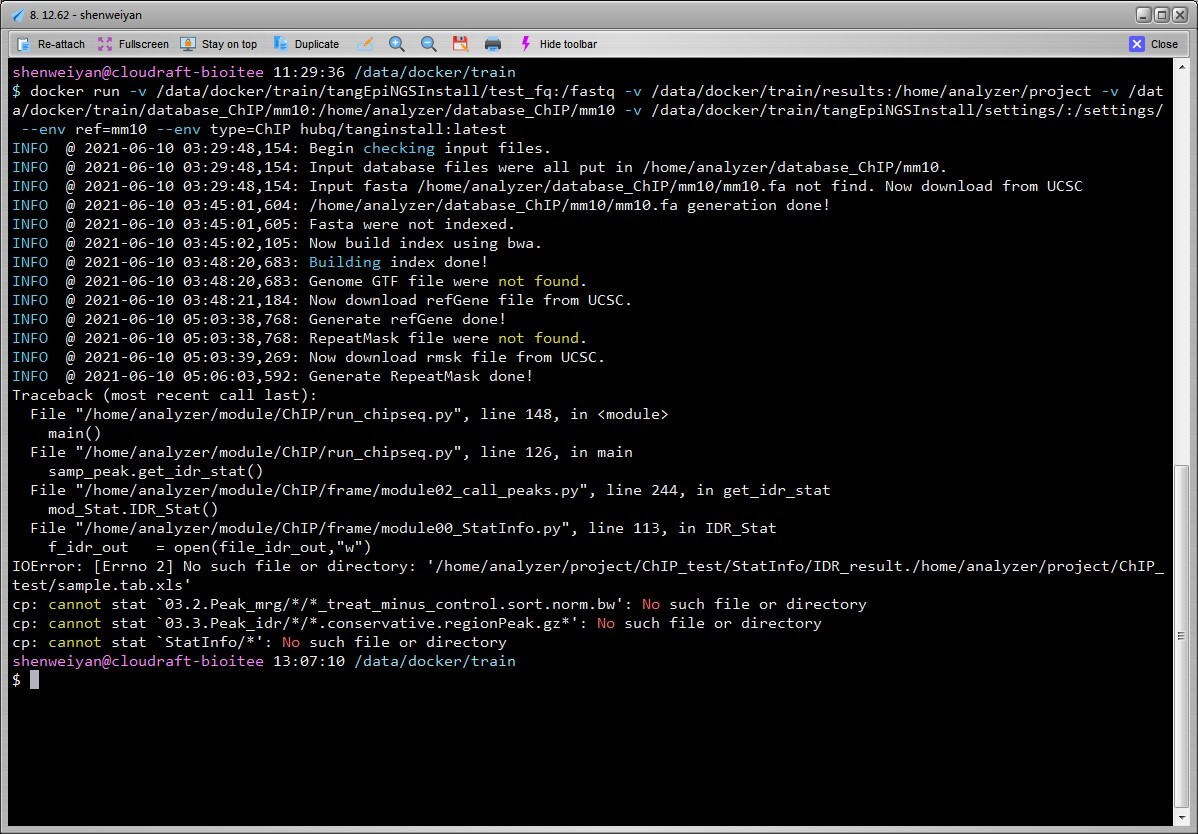
$ git clone https://github.com/shenweiyan/tangEpiNGSInstall.git$ tree.└── tangEpiNGSInstall├── Dockerfile├── README.md├── settings│ ├── run_chipseq.py│ ├── run_chipseq.sh│ ├── run_mRNA.py│ ├── run_mRNA.sh│ ├── scripts_chipseq.py│ └── scripts_mRNA.py├── src│ └── run_sample.sh├── test_fq│ ├── H3K4me3│ │ ├── test.1.fq.gz│ │ └── test.2.fq.gz│ ├── Input│ │ ├── test.1.fq.gz│ │ └── test.2.fq.gz│ └── sample.tab.xls└── test_fq_RNA├── SampleA1│ ├── test.1.fastq.gz│ └── test.2.fastq.gz└── sample.tab.xls8 directories, 17 files$ mkdir -p results database_ChIP/mm10$ chmod 777 results database_ChIP/mm10 # avoiding Permission issue$ tree.├── database_ChIP│ └── mm10├── results└── tangEpiNGSInstall├── Dockerfile├── README.md├── settings│ ├── run_chipseq.py│ ├── run_chipseq.sh│ ├── run_mRNA.py│ ├── run_mRNA.sh│ ├── scripts_chipseq.py│ └── scripts_mRNA.py├── src│ └── run_sample.sh├── test_fq│ ├── H3K4me3│ │ ├── test.1.fq.gz│ │ └── test.2.fq.gz│ ├── Input│ │ ├── test.1.fq.gz│ │ └── test.2.fq.gz│ └── sample.tab.xls└── test_fq_RNA├── SampleA1│ ├── test.1.fastq.gz│ └── test.2.fastq.gz└── sample.tab.xls11 directories, 17 files$ docker pull hubq/tanginstall:latest$ docker run -v /data/docker/train/tangEpiNGSInstall/test_fq:/fastq -v /data/docker/train/results:/home/analyzer/project -v /data/docker/train/database_ChIP/mm10:/home/analyzer/database_ChIP/mm10 -v /data/docker/train/tangEpiNGSInstall/settings/:/settings/ --env ref=mm10 --env type=ChIP hubq/tanginstall:latestINFO @ 2021-06-10 03:29:48,154: Begin checking input files.INFO @ 2021-06-10 03:29:48,154: Input database files were all put in /home/analyzer/database_ChIP/mm10.INFO @ 2021-06-10 03:29:48,154: Input fasta /home/analyzer/database_ChIP/mm10/mm10.fa not find. Now download from UCSCINFO @ 2021-06-10 03:45:01,604: /home/analyzer/database_ChIP/mm10/mm10.fa generation done!INFO @ 2021-06-10 03:45:01,605: Fasta were not indexed.INFO @ 2021-06-10 03:45:02,105: Now build index using bwa.INFO @ 2021-06-10 03:48:20,683: Building index done!INFO @ 2021-06-10 03:48:20,683: Genome GTF file were not found.INFO @ 2021-06-10 03:48:21,184: Now download refGene file from UCSC.INFO @ 2021-06-10 05:03:38,768: Generate refGene done!INFO @ 2021-06-10 05:03:38,768: RepeatMask file were not found.INFO @ 2021-06-10 05:03:39,269: Now download rmsk file from UCSC.INFO @ 2021-06-10 05:06:03,592: Generate RepeatMask done!Traceback (most recent call last):File "/home/analyzer/module/ChIP/run_chipseq.py", line 148, in <module>main()File "/home/analyzer/module/ChIP/run_chipseq.py", line 126, in mainsamp_peak.get_idr_stat()File "/home/analyzer/module/ChIP/frame/module02_call_peaks.py", line 244, in get_idr_statmod_Stat.IDR_Stat()File "/home/analyzer/module/ChIP/frame/module00_StatInfo.py", line 113, in IDR_Statf_idr_out = open(file_idr_out,"w")IOError: [Errno 2] No such file or directory: '/home/analyzer/project/ChIP_test/StatInfo/IDR_result./home/analyzer/project/ChIP_test/sample.tab.xls'cp: cannot stat `03.2.Peak_mrg/*/*_treat_minus_control.sort.norm.bw': No such file or directorycp: cannot stat `03.3.Peak_idr/*/*.conservative.regionPeak.gz*': No such file or directorycp: cannot stat `StatInfo/*': No such file or directory
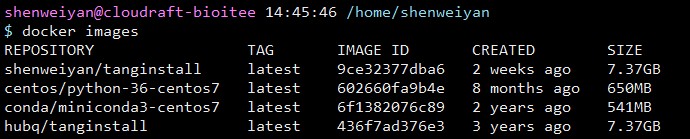
出于学习和折腾,针对这个问题,个人在 hubq/tanginstall:latest 的镜像基础上做了一点小调整,并重新打包成一个名为 shenweiyan/tanginstall:latest 的新镜像 push 到了 Docker Hub,抛砖引玉,供大家学习参考。
简单说一下这个镜像的几点细节。
整个镜像体积比较大,总共约 7.37GB,pull 下来可能比较慢。
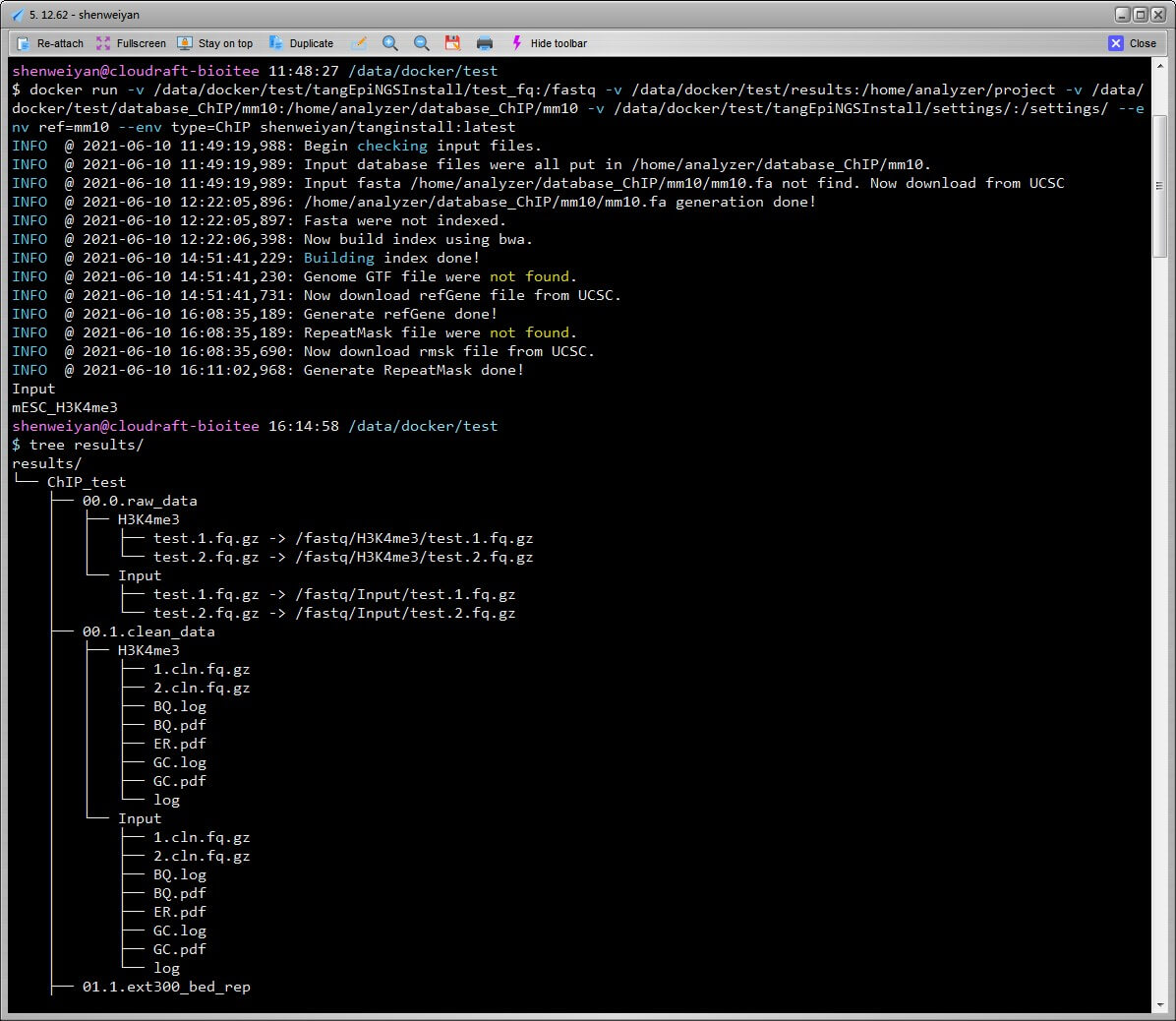
如果没有 ref(hg19/hg38 or mm9/mm10),镜像执行过程中会首先执行下载,然后拆分合并,建立 index。
- db01.DownloadRef.sh ``` $ cat db01.DownloadRef.sh ref=$1 dir_database=/home/analyzer/database_ChIP/$ref dir_path=/home/analyzer/module/ChIP
cd $dir_database
wget http://hgdownload.soe.ucsc.edu/goldenPath/${ref}/bigZips/chromFa.tar.gz
tar -zxvf $dir_database/chromFa.tar.gz
for i in {1..22} X Y M do cat $dir_database/chr$i.fa done >$dir_database/${ref}.fa && rm $dir_database/chr*fa
- db02.RefIndex.sh
$ cat db02.RefIndex.sh ref=$1 dir_database=/home/analyzer/database_ChIP/$ref bwa_exe=/software/install_packages/bwa-0.7.5a/bwa samtools_exe=/software/install_packages/samtools-0.1.18/samtools div_bins_exe=/home/analyzer/module/ChIP/bin/div_bins/bed_read
$samtools_exe faidx $dir_database/${ref}.fa
$bwa_exe index $dir_database/${ref}.fa
$dix_bins_exe -b 100 $dir_database/${ref}.fa.fai $dir_database/columns.100.bed $dix_bins_exe -b 1000 $dir_database/${ref}.fa.fai $dir_database/columns.1kb.bed
cut -f 1-2 $dir_database/${ref}.fa.fai >$dir_database/${ref}.fa.len
- db03.RefGene.sh
$ cat db03.RefGene.sh ref=$1 dir_database=/home/analyzer/database_ChIP/$ref bedtools_exe=/software/install_packages/bedtools2/bin/bedtools ucsc_dir=/software/install_packages/UCSC bin=/home/analyzer/module/ChIP/bin dir_path=/home/analyzer/module/ChIP
cd $dir_database wget http://hgdownload.soe.ucsc.edu/goldenPath/${ref}/database/refGene.txt.gz
remove chromosome fragments(unassembled).
for i in {1..22} X Y M do zcat $dir_database/refGene.txt.gz | grep -w chr$i done >$dir_database/tmp mv $dir_database/tmp $dir_database/refGene.txt
refGene.bed
cat $dir_database/refGene.txt |\ awk ‘{ tag=”noncoding”; if($4~/^NM/){tag=”protein_coding”}; OFS=”\t”; print $3,$5,$6,$2,$4,$10,$11,tag,$13 }’ /dev/stdin |\ python $bin/s03_genePred2bed.py /dev/stdin |\ $bedtools_exe sort -i /dev/stdin >$dir_database/refGene.bed &&\
region.Intragenic.bed
For novo lncRNA detection
$bin/find_ExonIntronIntergenic/find_ExonIntronIntergenic \ $dir_database/refGene.bed \ $dir_database/${ref}.fa.fai >$dir_database/pos.bed &&\
grep -v “Intergenic” $dir_database/pos.bed |\ awk ‘{OFS=” “;print $1,$2,$3,”Intragenic”}’ /dev/stdin \
>$dir_database/region.Intragenic.bed &&\
refGene.gtf
For mapping
zcat $dir_database/refGene.txt.gz |\ cut -f 2- |\ $ucsc_dir/genePredToGtf file stdin /dev/stdout |\ grep -w exon |\ $bedtools_exe sort -i /dev/stdin >$dir_database/refGene.gtf &&\ cat $dir_path/database/ERCC.gtf >>$dir_database/refGene.gtf
- db04.rmsk.sh
$ cat db04.rmsk.sh ref=$1 dir_database=/home/analyzer/database_ChIP/$ref bedtools_exe=/software/install_packages/bedtools2/bin/bedtools ucsc_dir=/software/install_packages/UCSC bin=/home/analyzer/module/ChIP/bin dir_path=/home/analyzer/module/ChIP
cd $dir_database wget http://hgdownload.soe.ucsc.edu/goldenPath/${ref}/database/rmsk.txt.gz
zcat $dir_database/rmsk.txt.gz |\ awk ‘{ OFS=”\t”; print $6,$7,$8,$2,”.”,”.”,”.”,”(“$9”)”,$10,$11,$12 “/“ $13,$14,$15,$16,$17 }’ /dev/stdin |\ tail -n +2 /dev/stdin >$dir_database/chrom.bed
for i in {1..22} X Y M do grep -w chr$i $dir_database/chrom.bed done >$dir_database/tmp mv $dir_database/tmp $dir_database/chrom.bed
$bedtools_exe sort -i $dir_database/chrom.bed >$dir_database/chrom.sort.bed
3. 为节省下载时间,建议事先准备好 ${ref}.fa,如果没有,也可以先下载好以下文件。
db01.DownloadRef.sh:
wget http://hgdownload.soe.ucsc.edu/goldenPath/${ref}/bigZips/chromFa.tar.gz
db03.RefGene.sh:
wget http://hgdownload.soe.ucsc.edu/goldenPath/${ref}/database/refGene.txt.gz
db04.rmsk.sh:
wget http://hgdownload.soe.ucsc.edu/goldenPath/${ref}/database/rmsk.txt.gz ```
- bwa index(db02.RefIndex.sh)非常耗时,个人一个4核16G配置的服务器也跑了大约2.5小时。

