import numpy as npimport torchimport torch.nn as nnimport torch.nn.functional as Ffrom torch_geometric.nn import GATConvimport torch_geometric.transforms as T
官方例子
import torchfrom torch_geometric.data import Dataedge_index = torch.tensor([[0, 1, 1, 2],[1, 0, 2, 1]], dtype=torch.long)x = torch.tensor([[-1], [0], [1]], dtype=torch.float)data = Data(x=x, edge_index=edge_index)
print(data.num_nodes)print(data.num_edges)print(data.num_node_features)print(data.is_directed())>>>341False
#转化成networkx格式from torch_geometric.utils import to_networkxdata_networkx = to_networkx(data, to_undirected=True)
使用plt绘图
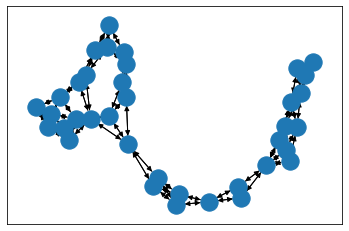
import matplotlib.pyplot as pltimport networkx as nxpos = nx.layout.spring_layout(data_networkx)# plt.figure(figsize=(16,12))nx.draw_networkx_nodes(data_networkx, pos)nx.draw_networkx_edges(data_networkx, pos,width=1,edge_color="black")plt.show()
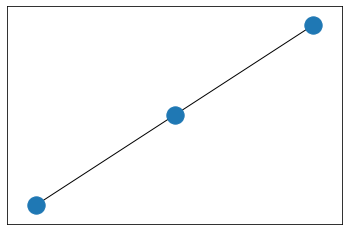
如果去掉data_networkx = to_networkx(data, to_undirected=True)中的to_undirected=True,结果如下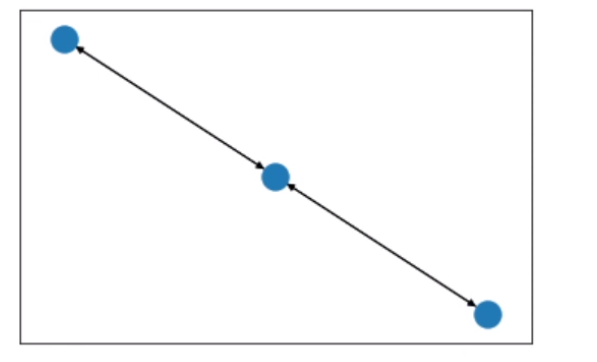
使用hvplot绘图
import matplotlib.pyplot as pltimport networkx as nximport hvplot.networkx as hvnxnode = hvnx.draw_networkx_nodes(data_networkx, pos)edge = hvnx.draw_networkx_edges(data_networkx, pos, arrowstyle='->', edge_width=2, colorbar=True)node * edge
封装绘图工具
def draw_with_nx(data_nx):import matplotlib.pyplot as pltimport networkx as nxpos = nx.layout.spring_layout(data_nx)nx.draw_networkx_nodes(data_nx, pos)nx.draw_networkx_edges(data_nx, pos,width=1,edge_color="black")plt.show()
def draw_with_hvplot(data_nx):import matplotlib.pyplot as pltimport networkx as nximport hvplot.networkx as hvnxpos = nx.layout.spring_layout(data_nx)node = hvnx.draw_networkx_nodes(data_nx, pos)edge = hvnx.draw_networkx_edges(data_nx, pos, arrowstyle='->', edge_width=2, colorbar=True)return node * edge
Networkx 内置的Graph Data Example
G_bar = nx.barabasi_albert_graph(n=100, m=3)draw_with_nx(G_bar)# nx.draw_kamada_kawai(G)
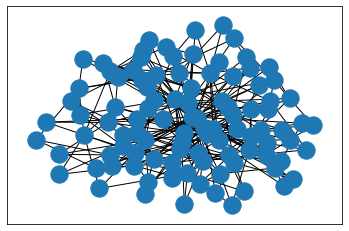
draw_with_hvplot(G_bar)
from torch_geometric import utilsdata = utils.from_networkx(G)
networkx转化成PyG格式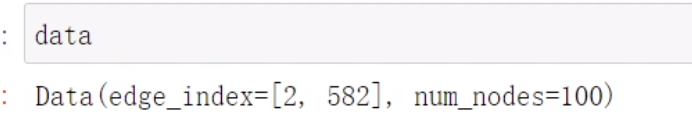 本质上只有edge_index起作用
本质上只有edge_index起作用
data.keys>>>['edge_index']
数据集
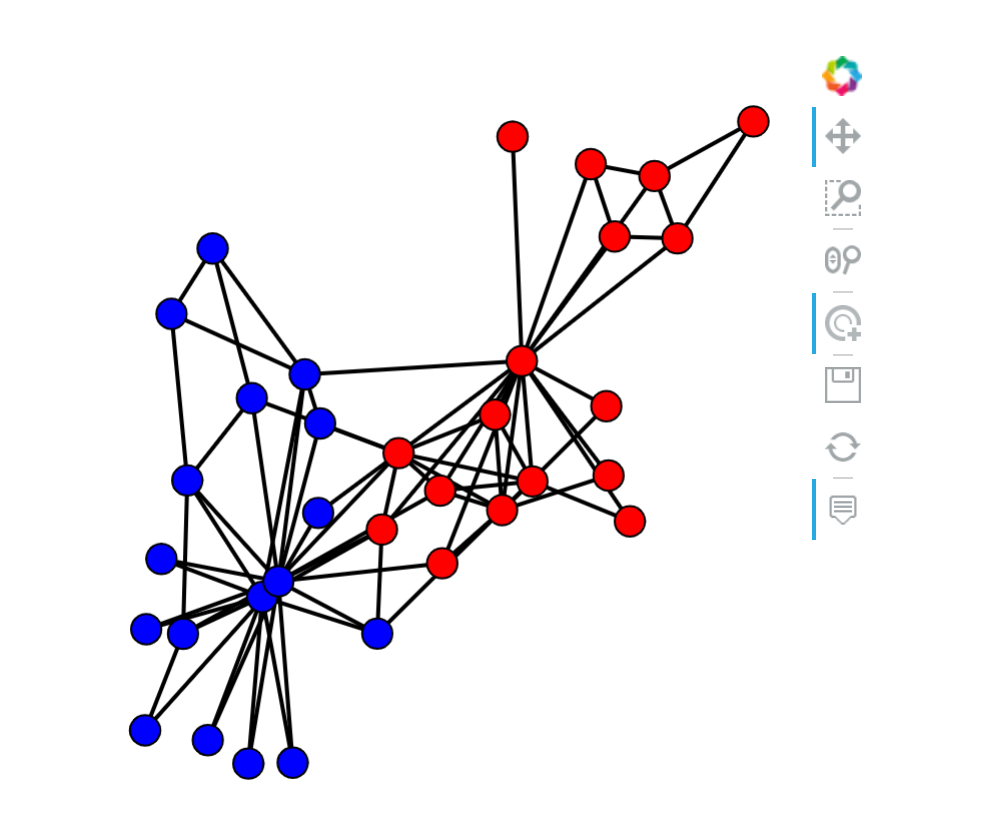
Networkx Karate Club空手道俱乐部
import networkx as nxG_karate = nx.karate_club_graph()data_karate = utils.from_networkx(G_karate)print(data_karate)>>>Data(club=[34], edge_index=[2, 156])
set(data_karate.club)>>>{'Mr. Hi', 'Officer'}
node_color = ["blue", "red"]node_label = np.array(list(G_karate.nodes))import matplotlib.pyplot as pltimport networkx as nximport hvplot.networkx as hvnxpos = nx.layout.spring_layout(G_karate)plt_nodes = []for i, c in enumerate(list(set(data_karate.club))):nodelist = [n for n in range(len(G_karate.nodes)) if G_karate.nodes[n]['club'] == c]plt = hvnx.draw_networkx_nodes(G_karate, pos, nodelist=list(nodelist), node_color=node_color[i])plt_nodes.append(plt)plt_edges = hvnx.draw_networkx_edges(G_karate, pos, arrowstyle='->', edge_width=2, colorbar=True)import functoolsimport operatorplt_edges * functools.reduce(operator.mul, plt_nodes)
Cora Dataset
from torch_geometric.datasets import Planetoidname_data = 'Cora'dataset_cora = Planetoid(root='/proj/data/', name=name_data)
print(type(dataset_cora))print(type(dataset_cora[0]))print(type(dataset_cora.data))print(dataset_cora[0])print(dataset_cora.data)print(dataset_cora.data.keys)>>><class 'torch_geometric.datasets.planetoid.Planetoid'><class 'torch_geometric.data.data.Data'><class 'torch_geometric.data.data.Data'>Data(edge_index=[2, 10556], test_mask=[2708], train_mask=[2708], val_mask=[2708], x=[2708, 1433], y=[2708])Data(edge_index=[2, 10556], test_mask=[2708], train_mask=[2708], val_mask=[2708], x=[2708, 1433], y=[2708])['x', 'edge_index', 'y', 'train_mask', 'val_mask', 'test_mask']
print(dataset_cora.data.y)>>>tensor([3, 4, 4, ..., 3, 3, 3])
Enzymes Dataset
from torch_geometric.datasets import TUDatasetfrom torch_geometric.data import DataLoaderdataset_enz = TUDataset('./data', name='ENZYMES', use_node_attr=True)
print(type(dataset_enz))print(type(dataset_enz[0]))print(type(dataset_enz.data))print(dataset_enz[0])print(dataset_enz.data)print(dataset_enz.data.keys)>>><class 'torch_geometric.datasets.tu_dataset.TUDataset'><class 'torch_geometric.data.data.Data'><class 'torch_geometric.data.data.Data'>Data(edge_index=[2, 168], x=[37, 21], y=[1])Data(edge_index=[2, 74564], x=[19580, 21], y=[600])['x', 'edge_index', 'y']
画出前3个graph
from torch_geometric.utils import to_networkxfor i in range(3):data_nx_enz = to_networkx(dataset_enz[i])draw_with_nx(data_nx_enz)
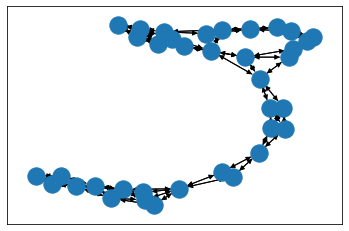
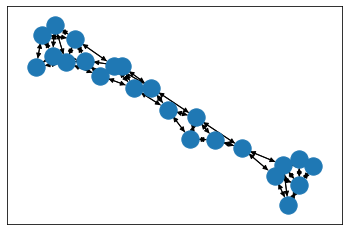
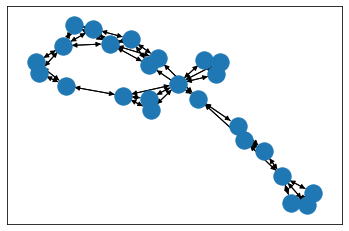
import matplotlib.pyplot as pltimport networkx as nxpos = nx.layout.spring_layout(data_nx_enz_0)# plt.figure(figsize=(16,12))nx.draw_networkx_nodes(data_nx_enz_0, pos)nx.draw_networkx_edges(data_nx_enz_0, pos,width=1,edge_color="black")plt.show()
Batch Graph
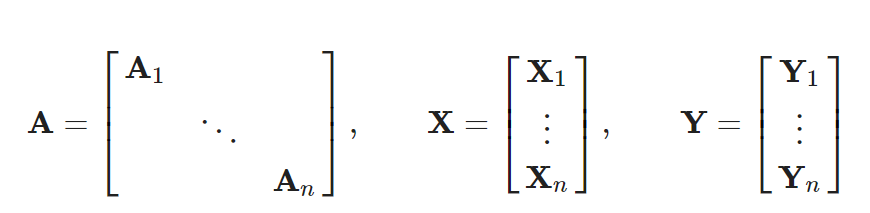
将Enzymes数据集的前10个graph组成一个batch
from torch_geometric.data import Batchb = Batch.from_data_list(dataset_enz[:10])b>>>Batch(batch=[325], edge_index=[2, 1202], ptr=[11], x=[325, 21], y=[10])b.edge_index[0]>>>tensor([ 0, 0, 0, ..., 324, 324, 324])
将整个数据集以batch展示.
观察到batch大小的节点大小是一样的。这里的batch是说明每个x出现在哪个graph里
loader = DataLoader(dataset_enz, batch_size=64, shuffle=True)for batch in loader:print(batch)>>>Batch(batch=[1993], edge_index=[2, 7806], ptr=[65], x=[1993, 21], y=[64])Batch(batch=[2098], edge_index=[2, 8016], ptr=[65], x=[2098, 21], y=[64])Batch(batch=[2135], edge_index=[2, 7752], ptr=[65], x=[2135, 21], y=[64])Batch(batch=[1977], edge_index=[2, 7634], ptr=[65], x=[1977, 21], y=[64])Batch(batch=[1946], edge_index=[2, 7482], ptr=[65], x=[1946, 21], y=[64])Batch(batch=[2093], edge_index=[2, 7986], ptr=[65], x=[2093, 21], y=[64])Batch(batch=[2239], edge_index=[2, 8514], ptr=[65], x=[2239, 21], y=[64])Batch(batch=[2053], edge_index=[2, 8008], ptr=[65], x=[2053, 21], y=[64])Batch(batch=[2219], edge_index=[2, 8462], ptr=[65], x=[2219, 21], y=[64])Batch(batch=[827], edge_index=[2, 2904], ptr=[25], x=[827, 21], y=[24])

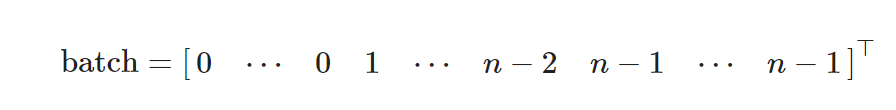
Two Graph Batch Example
import torchfrom torch_geometric.data import Dataedge_index = torch.tensor([[0, 1, 1, 2],[1, 0, 2, 1]], dtype=torch.long)x = torch.tensor([[-1], [0], [1]], dtype=torch.float)data1 = Data(x=x, edge_index=edge_index)data2 = Data(x=x, edge_index=edge_index)
from torch_geometric.data import Batchg_2 = Batch.from_data_list([data1, data2])
draw_with_nx(to_networkx(g_2, to_undirected=True))

draw_with_hvplot(to_networkx(g_2, to_undirected=True))
g_2.batch>>>tensor([0, 0, 0, 1, 1, 1])
g_2.edge_index>>>tensor([[0, 1, 1, 2, 3, 4, 4, 5],[1, 0, 2, 1, 4, 3, 5, 4]])
g_2_nx = to_networkx(g_2, to_undirected=True)g_2_nx.is_directed()>>>Falseprint(nx.number_connected_components(g_2_nx))>>>2
 可交互式的
可交互式的