
- mean 代表平均值,se 代表标准差,worst 代表最大值(3 个最大值的平均值)
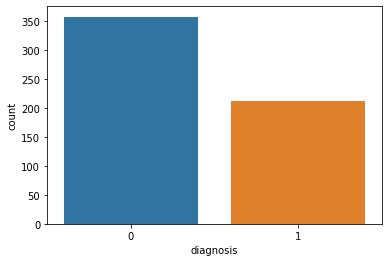
- 实际上是 10 个特征值(radius、texture、perimeter、area、smoothness、compactness、concavity、concave points、symmetry 和 fractal_dimension_mean)的 3 个维度,平均、标准差和最大值。这些特征值都保留了 4 位数字。字段中没有缺失的值。在 569 个患者中,一共有 357 个是良性,212 个是恶性。
import pandas as pdimport matplotlib.pyplot as plt# 画图import seaborn as snsfrom sklearn.model_selection import train_test_split#训练集与测试集from sklearn import svmfrom sklearn import metrics #?from sklearn.preprocessing import StandardScaler#标准分
data=pd.read_csv('data.csv')data.info()
<class 'pandas.core.frame.DataFrame'>RangeIndex: 569 entries, 0 to 568Data columns (total 32 columns):id 569 non-null int64diagnosis 569 non-null objectradius_mean 569 non-null float64texture_mean 569 non-null float64perimeter_mean 569 non-null float64area_mean 569 non-null float64smoothness_mean 569 non-null float64compactness_mean 569 non-null float64concavity_mean 569 non-null float64concave points_mean 569 non-null float64symmetry_mean 569 non-null float64fractal_dimension_mean 569 non-null float64radius_se 569 non-null float64texture_se 569 non-null float64perimeter_se 569 non-null float64area_se 569 non-null float64smoothness_se 569 non-null float64compactness_se 569 non-null float64concavity_se 569 non-null float64concave points_se 569 non-null float64symmetry_se 569 non-null float64fractal_dimension_se 569 non-null float64radius_worst 569 non-null float64texture_worst 569 non-null float64perimeter_worst 569 non-null float64area_worst 569 non-null float64smoothness_worst 569 non-null float64compactness_worst 569 non-null float64concavity_worst 569 non-null float64concave points_worst 569 non-null float64symmetry_worst 569 non-null float64fractal_dimension_worst 569 non-null float64dtypes: float64(30), int64(1), object(1)memory usage: 142.3+ KB
pd.set_option('display.max_columns',None)# 显示完整列
data.describe()
|
id |
radius_mean |
texture_mean |
perimeter_mean |
area_mean |
smoothness_mean |
compactness_mean |
concavity_mean |
concave points_mean |
symmetry_mean |
fractal_dimension_mean |
radius_se |
texture_se |
perimeter_se |
area_se |
smoothness_se |
compactness_se |
concavity_se |
concave points_se |
symmetry_se |
fractal_dimension_se |
radius_worst |
texture_worst |
perimeter_worst |
area_worst |
smoothness_worst |
compactness_worst |
concavity_worst |
concave points_worst |
symmetry_worst |
fractal_dimension_worst |
| count |
5.690000e+02 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
569.000000 |
| mean |
3.037183e+07 |
14.127292 |
19.289649 |
91.969033 |
654.889104 |
0.096360 |
0.104341 |
0.088799 |
0.048919 |
0.181162 |
0.062798 |
0.405172 |
1.216853 |
2.866059 |
40.337079 |
0.007041 |
0.025478 |
0.031894 |
0.011796 |
0.020542 |
0.003795 |
16.269190 |
25.677223 |
107.261213 |
880.583128 |
0.132369 |
0.254265 |
0.272188 |
0.114606 |
0.290076 |
0.083946 |
| std |
1.250206e+08 |
3.524049 |
4.301036 |
24.298981 |
351.914129 |
0.014064 |
0.052813 |
0.079720 |
0.038803 |
0.027414 |
0.007060 |
0.277313 |
0.551648 |
2.021855 |
45.491006 |
0.003003 |
0.017908 |
0.030186 |
0.006170 |
0.008266 |
0.002646 |
4.833242 |
6.146258 |
33.602542 |
569.356993 |
0.022832 |
0.157336 |
0.208624 |
0.065732 |
0.061867 |
0.018061 |
| min |
8.670000e+03 |
6.981000 |
9.710000 |
43.790000 |
143.500000 |
0.052630 |
0.019380 |
0.000000 |
0.000000 |
0.106000 |
0.049960 |
0.111500 |
0.360200 |
0.757000 |
6.802000 |
0.001713 |
0.002252 |
0.000000 |
0.000000 |
0.007882 |
0.000895 |
7.930000 |
12.020000 |
50.410000 |
185.200000 |
0.071170 |
0.027290 |
0.000000 |
0.000000 |
0.156500 |
0.055040 |
| 25% |
8.692180e+05 |
11.700000 |
16.170000 |
75.170000 |
420.300000 |
0.086370 |
0.064920 |
0.029560 |
0.020310 |
0.161900 |
0.057700 |
0.232400 |
0.833900 |
1.606000 |
17.850000 |
0.005169 |
0.013080 |
0.015090 |
0.007638 |
0.015160 |
0.002248 |
13.010000 |
21.080000 |
84.110000 |
515.300000 |
0.116600 |
0.147200 |
0.114500 |
0.064930 |
0.250400 |
0.071460 |
| 50% |
9.060240e+05 |
13.370000 |
18.840000 |
86.240000 |
551.100000 |
0.095870 |
0.092630 |
0.061540 |
0.033500 |
0.179200 |
0.061540 |
0.324200 |
1.108000 |
2.287000 |
24.530000 |
0.006380 |
0.020450 |
0.025890 |
0.010930 |
0.018730 |
0.003187 |
14.970000 |
25.410000 |
97.660000 |
686.500000 |
0.131300 |
0.211900 |
0.226700 |
0.099930 |
0.282200 |
0.080040 |
| 75% |
8.813129e+06 |
15.780000 |
21.800000 |
104.100000 |
782.700000 |
0.105300 |
0.130400 |
0.130700 |
0.074000 |
0.195700 |
0.066120 |
0.478900 |
1.474000 |
3.357000 |
45.190000 |
0.008146 |
0.032450 |
0.042050 |
0.014710 |
0.023480 |
0.004558 |
18.790000 |
29.720000 |
125.400000 |
1084.000000 |
0.146000 |
0.339100 |
0.382900 |
0.161400 |
0.317900 |
0.092080 |
| max |
9.113205e+08 |
28.110000 |
39.280000 |
188.500000 |
2501.000000 |
0.163400 |
0.345400 |
0.426800 |
0.201200 |
0.304000 |
0.097440 |
2.873000 |
4.885000 |
21.980000 |
542.200000 |
0.031130 |
0.135400 |
0.396000 |
0.052790 |
0.078950 |
0.029840 |
36.040000 |
49.540000 |
251.200000 |
4254.000000 |
0.222600 |
1.058000 |
1.252000 |
0.291000 |
0.663800 |
0.207500 |
# 字段分成3组
features_mean=list(data.columns[2:12])
features_se=list(data.columns[12:22])
features_worst=list(data.columns[22:32])
data.drop('id',axis=1,inplace=True)
data['diagnosis']=data['diagnosis'].map({'M':1,'B':0})
# 可视化
sns.countplot(data['diagnosis'],label="Count")
plt.show()
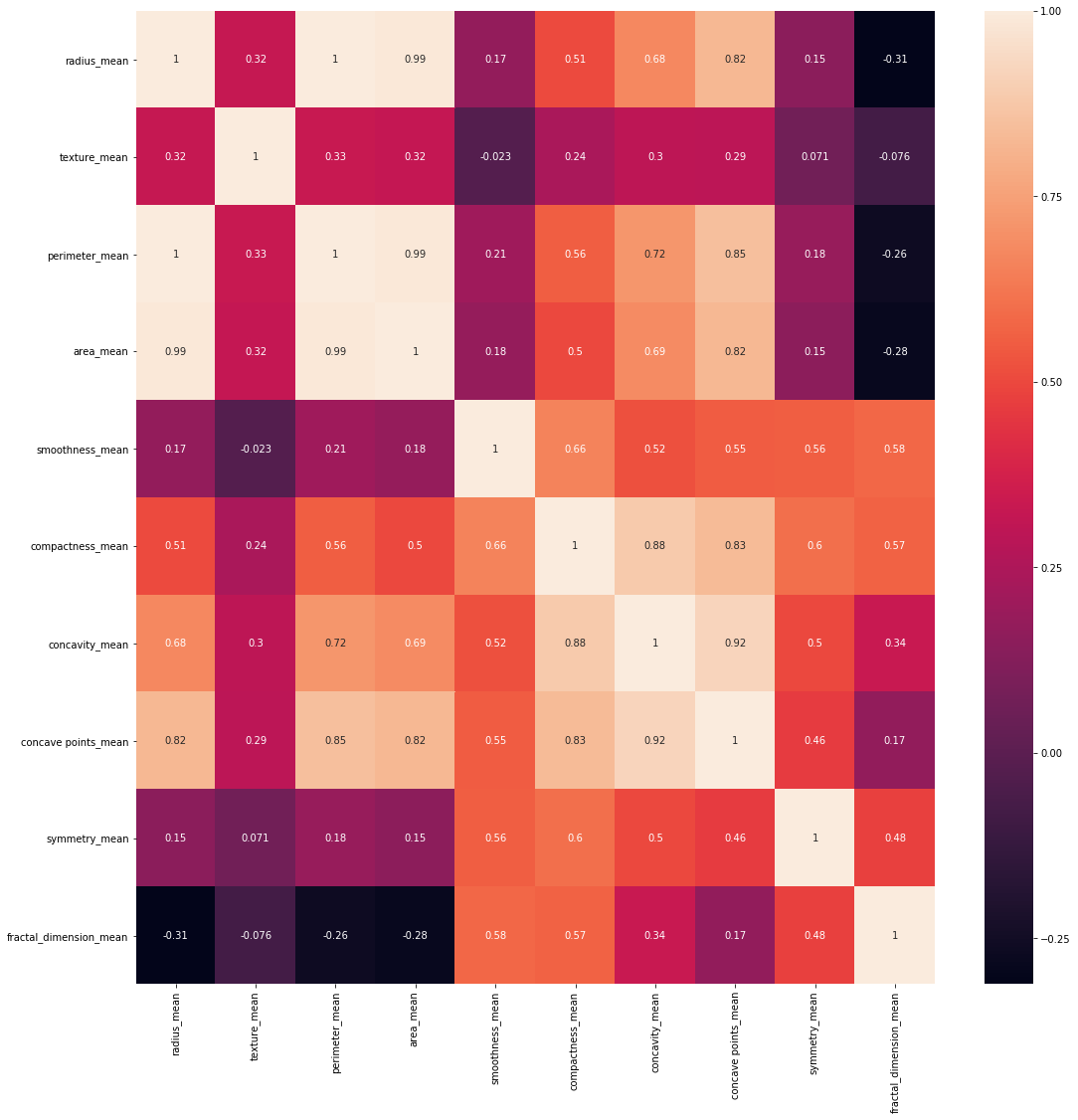
#利用热力图呈现与均值的关系
corr=data[features_mean].corr()#字段间的相关性(计算pearson相关系数)
plt.figure(figsize=(18,18))
sns.heatmap(corr,annot=True)
<matplotlib.axes._subplots.AxesSubplot at 0x1a31c99828>
features_remain = ['radius_mean','texture_mean', 'smoothness_mean','compactness_mean','symmetry_mean', 'fractal_dimension_mean']
train,test=train_test_split(data,test_size=0.3)
train_x=train[features_remain]
train_y=train['diagnosis']
test_x=train[features_remain]
test_y=train['diagnosis']
ss=StandardScaler()
train_x=ss.fit_transform(train_x)
test_x=ss.fit_transform(test_x)
model1=svm.SVC()
model1.fit(train_x,train_y)
SVC(C=1.0, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape='ovr', degree=3, gamma='auto_deprecated',
kernel='rbf', max_iter=-1, probability=False, random_state=None,
shrinking=True, tol=0.001, verbose=False)
prediction=model1.predict(test_x)
score=metrics.accuracy_score(prediction,test_y)
print(score)
0.9472361809045227
model2=svm.LinearSVC()
model2.fit(train_x,train_y)
prediction=model2.predict(test_x)
score=metrics.accuracy_score(prediction,test_y)
print(score)
0.9346733668341709
数据集
data.csv




