参考:https://cloud.tencent.com/developer/article/1814104 参考:https://cloud.tencent.com/developer/article/1842793?from=article.detail.1814104 注:仅作为笔记,不做商用。
harmony原理
Harmony需要输入低维空间的坐标值(embedding),一般使用PCA的降维结果。Harmony导入PCA的降维数据后,会采用soft k-means clustering算法将细胞聚类。常用的聚类算法仅考虑细胞在低维空间的距离,但是soft clustering算法会考虑我们提供的校正因素。这就好比我们的高考加分制度,小明高考成绩本来达不到A大学的录取分数线,但是他有一项省级竞赛一等奖加10分就够线了。同样的道理,细胞c2距离cluster1有点远,本来不能算作cluster1的一份子;但是c2和cluster1的细胞来自不同的数据集,因为我们期望不同的数据集融合,所以破例让它加入cluster1了。聚类之后先计算每个cluster内各个数据集的细胞的中心点,然后根据这些中心点计算各个cluster的中心点。最后通过算法让cluster内的细胞向中心聚集,实在收敛不了的离群细胞就过滤掉。调整之后的数据重复:聚类—计算cluster中心点—收敛细胞—聚类的过程,不断迭代直至聚类效果趋于稳定。
官方图解
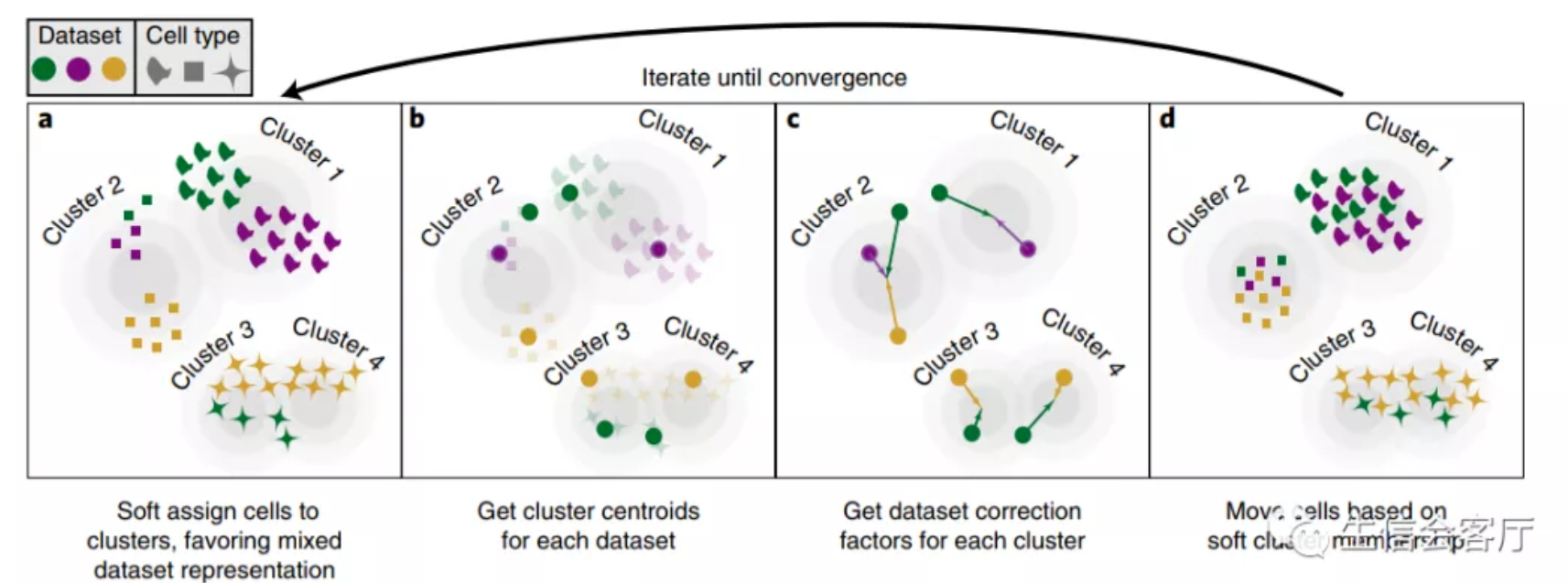
Overview of Harmony algorithm. PCA embeds cells into a space with reduced dimensionality. Harmony accepts the cell coordinates in this reduced space and runs an iterative algorithm to adjust for dataset specific effects.
- a, Harmony uses fuzzy clustering to assign each cell to multiple clusters, while a penalty term ensures that the diversity of datasets within each cluster is maximized.
- b, Harmony calculates a global centroid for each cluster, as well as dataset-specific centroids for each cluster.
- c, Within each cluster, Harmony calculates a correction factor for each dataset based on the centroids.
- d, Finally, Harmony corrects each cell with a cell-specific factor: a linear combination of dataset correction factors weighted by the cell’s soft cluster assignments made in step a.
实战
运行harmony仅需要一行代码,group.by.vars参数代表整合哪些组别的样本。library(harmony)seuratObj <- RunHarmony(sce, group.by.vars = "orig.ident")

