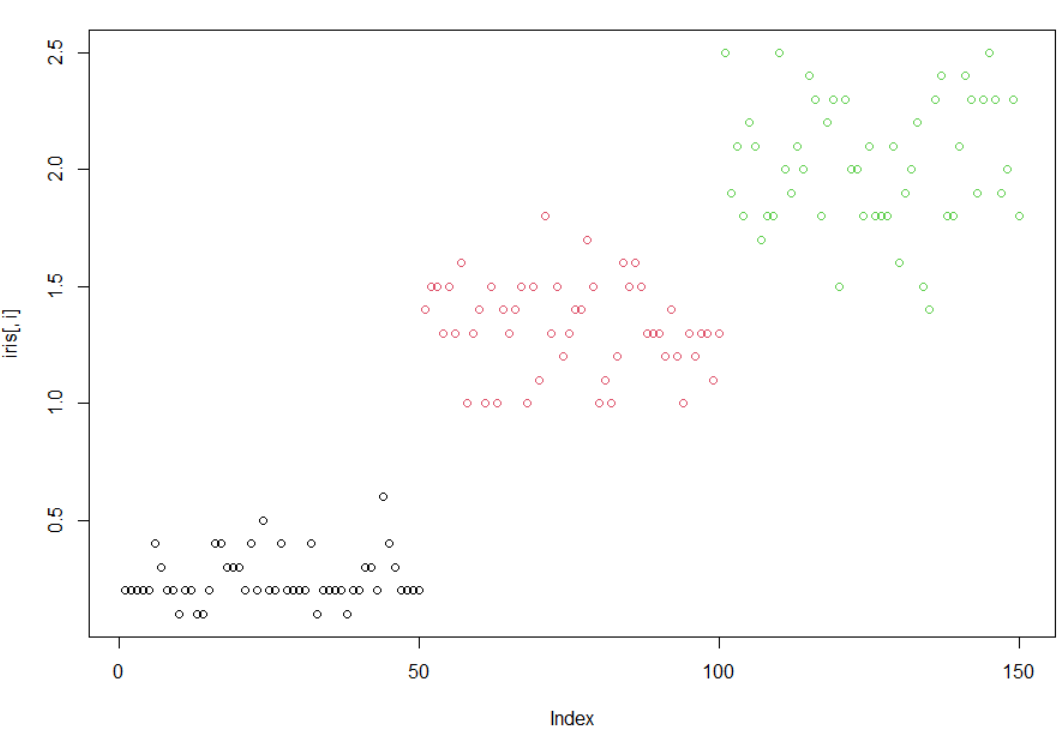
1使用循环,对iris的1到4列分别画点图(plot)
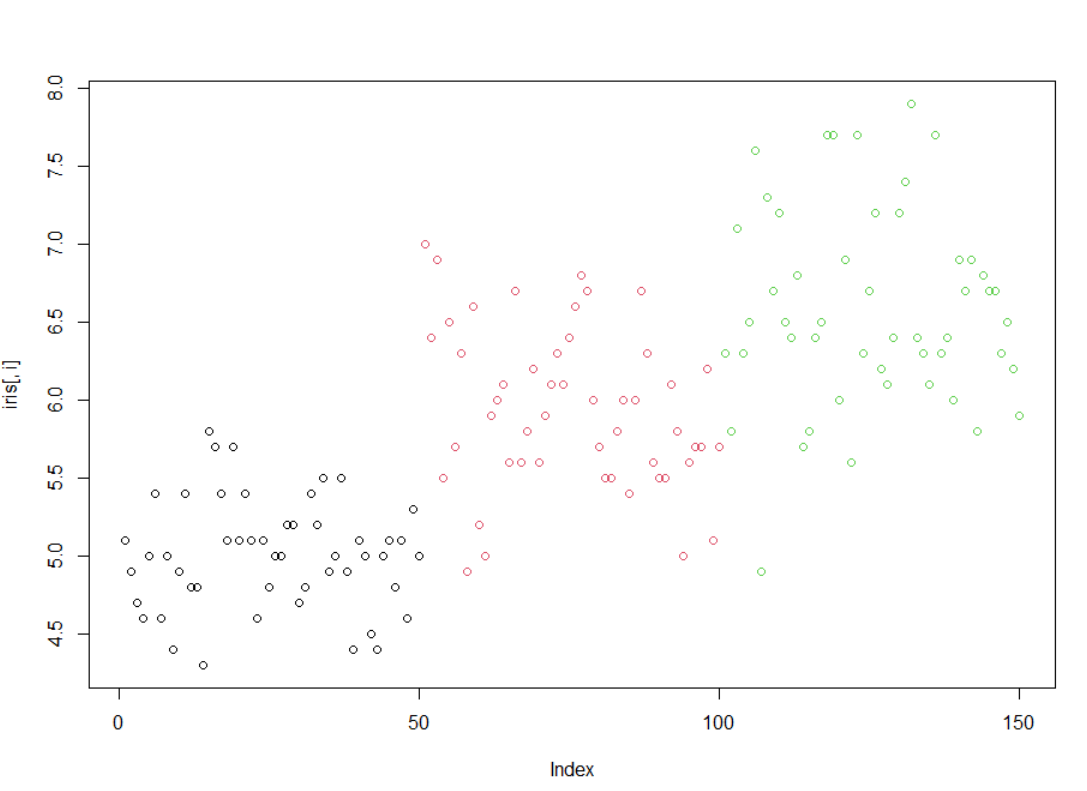
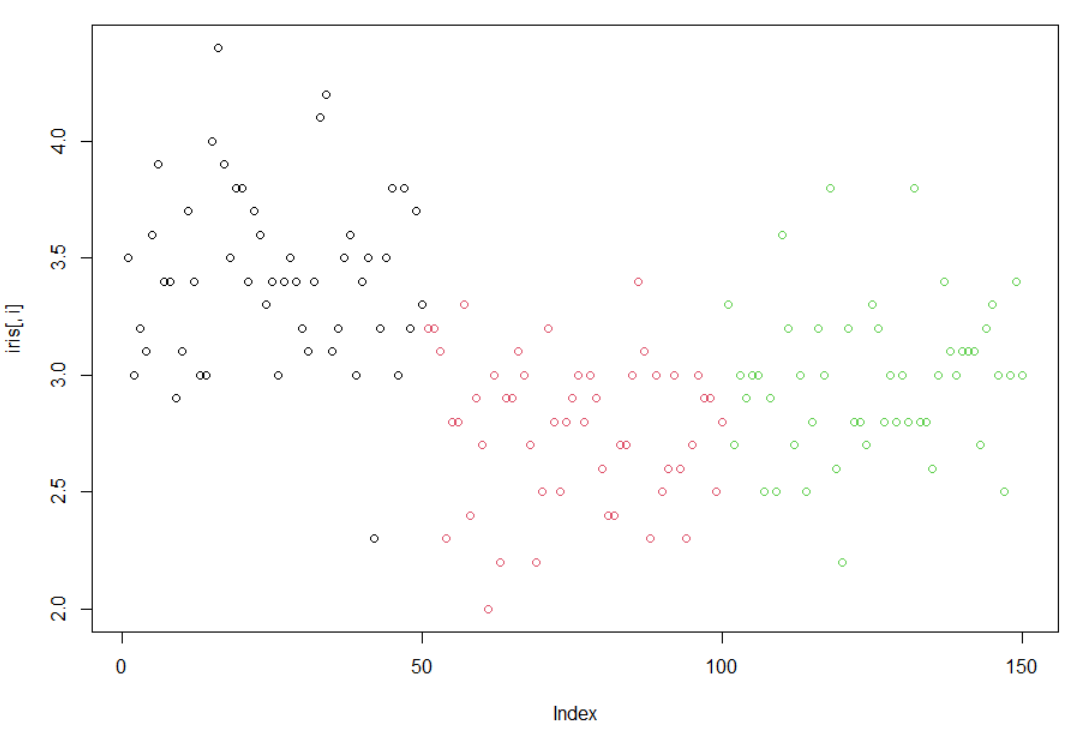
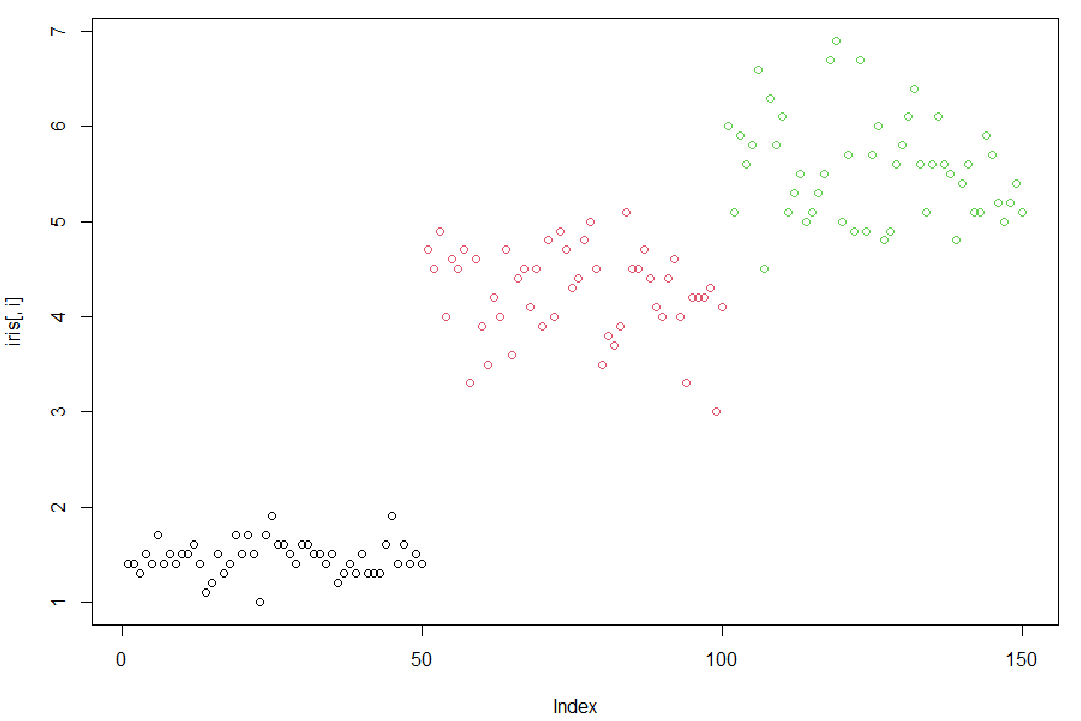
> irisSepal.Length Sepal.Width Petal.Length Petal.Width Species1 5.1 3.5 1.4 0.2 setosa2 4.9 3.0 1.4 0.2 setosa3 4.7 3.2 1.3 0.2 setosa4 4.6 3.1 1.5 0.2 setosa5 5.0 3.6 1.4 0.2 setosa6 5.4 3.9 1.7 0.4 setosa7 4.6 3.4 1.4 0.3 setosa8 5.0 3.4 1.5 0.2 setosa9 4.4 2.9 1.4 0.2 setosa10 4.9 3.1 1.5 0.1 setosa11 5.4 3.7 1.5 0.2 setosa12 4.8 3.4 1.6 0.2 setosa13 4.8 3.0 1.4 0.1 setosa14 4.3 3.0 1.1 0.1 setosa15 5.8 4.0 1.2 0.2 setosa16 5.7 4.4 1.5 0.4 setosa17 5.4 3.9 1.3 0.4 setosa18 5.1 3.5 1.4 0.3 setosa19 5.7 3.8 1.7 0.3 setosa20 5.1 3.8 1.5 0.3 setosa21 5.4 3.4 1.7 0.2 setosa22 5.1 3.7 1.5 0.4 setosa23 4.6 3.6 1.0 0.2 setosa24 5.1 3.3 1.7 0.5 setosa25 4.8 3.4 1.9 0.2 setosa26 5.0 3.0 1.6 0.2 setosa27 5.0 3.4 1.6 0.4 setosa28 5.2 3.5 1.5 0.2 setosa29 5.2 3.4 1.4 0.2 setosa30 4.7 3.2 1.6 0.2 setosa31 4.8 3.1 1.6 0.2 setosa32 5.4 3.4 1.5 0.4 setosa33 5.2 4.1 1.5 0.1 setosa34 5.5 4.2 1.4 0.2 setosa35 4.9 3.1 1.5 0.2 setosa36 5.0 3.2 1.2 0.2 setosa37 5.5 3.5 1.3 0.2 setosa38 4.9 3.6 1.4 0.1 setosa39 4.4 3.0 1.3 0.2 setosa40 5.1 3.4 1.5 0.2 setosa41 5.0 3.5 1.3 0.3 setosa42 4.5 2.3 1.3 0.3 setosa43 4.4 3.2 1.3 0.2 setosa44 5.0 3.5 1.6 0.6 setosa45 5.1 3.8 1.9 0.4 setosa46 4.8 3.0 1.4 0.3 setosa47 5.1 3.8 1.6 0.2 setosa48 4.6 3.2 1.4 0.2 setosa49 5.3 3.7 1.5 0.2 setosa50 5.0 3.3 1.4 0.2 setosa51 7.0 3.2 4.7 1.4 versicolor52 6.4 3.2 4.5 1.5 versicolor53 6.9 3.1 4.9 1.5 versicolor54 5.5 2.3 4.0 1.3 versicolor55 6.5 2.8 4.6 1.5 versicolor56 5.7 2.8 4.5 1.3 versicolor57 6.3 3.3 4.7 1.6 versicolor58 4.9 2.4 3.3 1.0 versicolor59 6.6 2.9 4.6 1.3 versicolor60 5.2 2.7 3.9 1.4 versicolor61 5.0 2.0 3.5 1.0 versicolor62 5.9 3.0 4.2 1.5 versicolor63 6.0 2.2 4.0 1.0 versicolor64 6.1 2.9 4.7 1.4 versicolor65 5.6 2.9 3.6 1.3 versicolor66 6.7 3.1 4.4 1.4 versicolor67 5.6 3.0 4.5 1.5 versicolor68 5.8 2.7 4.1 1.0 versicolor69 6.2 2.2 4.5 1.5 versicolor70 5.6 2.5 3.9 1.1 versicolor71 5.9 3.2 4.8 1.8 versicolor72 6.1 2.8 4.0 1.3 versicolor73 6.3 2.5 4.9 1.5 versicolor74 6.1 2.8 4.7 1.2 versicolor75 6.4 2.9 4.3 1.3 versicolor76 6.6 3.0 4.4 1.4 versicolor77 6.8 2.8 4.8 1.4 versicolor78 6.7 3.0 5.0 1.7 versicolor79 6.0 2.9 4.5 1.5 versicolor80 5.7 2.6 3.5 1.0 versicolor81 5.5 2.4 3.8 1.1 versicolor82 5.5 2.4 3.7 1.0 versicolor83 5.8 2.7 3.9 1.2 versicolor84 6.0 2.7 5.1 1.6 versicolor85 5.4 3.0 4.5 1.5 versicolor86 6.0 3.4 4.5 1.6 versicolor87 6.7 3.1 4.7 1.5 versicolor88 6.3 2.3 4.4 1.3 versicolor89 5.6 3.0 4.1 1.3 versicolor90 5.5 2.5 4.0 1.3 versicolor91 5.5 2.6 4.4 1.2 versicolor92 6.1 3.0 4.6 1.4 versicolor93 5.8 2.6 4.0 1.2 versicolor94 5.0 2.3 3.3 1.0 versicolor95 5.6 2.7 4.2 1.3 versicolor96 5.7 3.0 4.2 1.2 versicolor97 5.7 2.9 4.2 1.3 versicolor98 6.2 2.9 4.3 1.3 versicolor99 5.1 2.5 3.0 1.1 versicolor100 5.7 2.8 4.1 1.3 versicolor101 6.3 3.3 6.0 2.5 virginica102 5.8 2.7 5.1 1.9 virginica103 7.1 3.0 5.9 2.1 virginica104 6.3 2.9 5.6 1.8 virginica105 6.5 3.0 5.8 2.2 virginica106 7.6 3.0 6.6 2.1 virginica107 4.9 2.5 4.5 1.7 virginica108 7.3 2.9 6.3 1.8 virginica109 6.7 2.5 5.8 1.8 virginica110 7.2 3.6 6.1 2.5 virginica111 6.5 3.2 5.1 2.0 virginica112 6.4 2.7 5.3 1.9 virginica113 6.8 3.0 5.5 2.1 virginica114 5.7 2.5 5.0 2.0 virginica115 5.8 2.8 5.1 2.4 virginica116 6.4 3.2 5.3 2.3 virginica117 6.5 3.0 5.5 1.8 virginica118 7.7 3.8 6.7 2.2 virginica119 7.7 2.6 6.9 2.3 virginica120 6.0 2.2 5.0 1.5 virginica121 6.9 3.2 5.7 2.3 virginica122 5.6 2.8 4.9 2.0 virginica123 7.7 2.8 6.7 2.0 virginica124 6.3 2.7 4.9 1.8 virginica125 6.7 3.3 5.7 2.1 virginica126 7.2 3.2 6.0 1.8 virginica127 6.2 2.8 4.8 1.8 virginica128 6.1 3.0 4.9 1.8 virginica129 6.4 2.8 5.6 2.1 virginica130 7.2 3.0 5.8 1.6 virginica131 7.4 2.8 6.1 1.9 virginica132 7.9 3.8 6.4 2.0 virginica133 6.4 2.8 5.6 2.2 virginica134 6.3 2.8 5.1 1.5 virginica135 6.1 2.6 5.6 1.4 virginica136 7.7 3.0 6.1 2.3 virginica137 6.3 3.4 5.6 2.4 virginica138 6.4 3.1 5.5 1.8 virginica139 6.0 3.0 4.8 1.8 virginica140 6.9 3.1 5.4 2.1 virginica141 6.7 3.1 5.6 2.4 virginica142 6.9 3.1 5.1 2.3 virginica143 5.8 2.7 5.1 1.9 virginica144 6.8 3.2 5.9 2.3 virginica145 6.7 3.3 5.7 2.5 virginica146 6.7 3.0 5.2 2.3 virginica147 6.3 2.5 5.0 1.9 virginica148 6.5 3.0 5.2 2.0 virginica149 6.2 3.4 5.4 2.3 virginica150 5.9 3.0 5.1 1.8 virginica> x=1:4> x[1] 1 2 3 4> for (i in x){+ plot(iris[,i],col=iris[,5])+ }
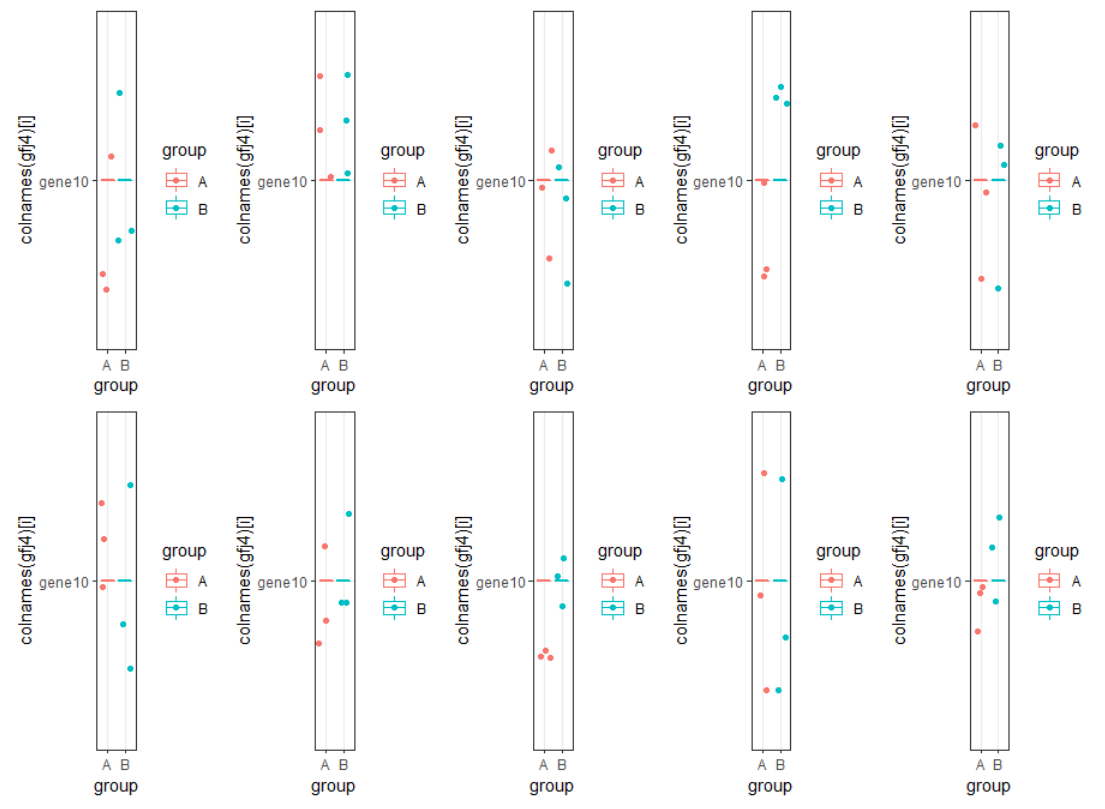
2生成一个随机数(rnorm)组成的10行6列的矩阵,列名为sample1,sample2….sample6,行名为gene1,gene2…gene10,分组为sample1、2、3属于A组,sample4,5、6属于B组。用循环对每个基因画ggplot2箱线图,并拼图
> gfj = matrix(rnorm(60),ncol=6);gfj[,1] [,2] [,3] [,4] [,5] [,6][1,] -0.31268804 0.9324936 -1.182418259 1.7039155 -0.7824128 -0.2051285[2,] -0.61609178 1.4331777 -0.190183535 -0.4211536 0.6690160 0.4631609[3,] -0.40235005 1.3724106 1.550675136 -2.7163350 0.7792040 -0.2501707[4,] 0.81463196 -0.2542318 -1.345814036 0.8897545 -0.2907730 -1.0246765[5,] -0.03485606 -0.6182081 -0.811909210 1.0200308 -0.9567138 0.3179821[6,] -0.70287745 -1.4913486 -0.006954597 -0.6774349 1.7549044 -0.4808167[7,] 0.09638077 0.5397555 1.588257406 -0.6570834 -0.3611543 0.2163203[8,] 0.66215689 1.4717037 1.450286698 -0.8983272 0.4830548 -2.0012366[9,] 0.20817293 1.4901270 0.598326457 -0.9547770 0.7461906 -1.0561868[10,] 0.40870489 -1.8879957 0.715517402 0.2257603 0.9773011 -0.4206816> rownames(gfj)=paste0(rep("gene",times=10),1:10);gfj[,1] [,2] [,3] [,4] [,5] [,6]gene1 -0.31268804 0.9324936 -1.182418259 1.7039155 -0.7824128 -0.2051285gene2 -0.61609178 1.4331777 -0.190183535 -0.4211536 0.6690160 0.4631609gene3 -0.40235005 1.3724106 1.550675136 -2.7163350 0.7792040 -0.2501707gene4 0.81463196 -0.2542318 -1.345814036 0.8897545 -0.2907730 -1.0246765gene5 -0.03485606 -0.6182081 -0.811909210 1.0200308 -0.9567138 0.3179821gene6 -0.70287745 -1.4913486 -0.006954597 -0.6774349 1.7549044 -0.4808167gene7 0.09638077 0.5397555 1.588257406 -0.6570834 -0.3611543 0.2163203gene8 0.66215689 1.4717037 1.450286698 -0.8983272 0.4830548 -2.0012366gene9 0.20817293 1.4901270 0.598326457 -0.9547770 0.7461906 -1.0561868gene10 0.40870489 -1.8879957 0.715517402 0.2257603 0.9773011 -0.4206816> colnames(gfj)=paste0(rep("sample",times=6),1:6);gfjsample1 sample2 sample3 sample4 sample5 sample6gene1 -0.31268804 0.9324936 -1.182418259 1.7039155 -0.7824128 -0.2051285gene2 -0.61609178 1.4331777 -0.190183535 -0.4211536 0.6690160 0.4631609gene3 -0.40235005 1.3724106 1.550675136 -2.7163350 0.7792040 -0.2501707gene4 0.81463196 -0.2542318 -1.345814036 0.8897545 -0.2907730 -1.0246765gene5 -0.03485606 -0.6182081 -0.811909210 1.0200308 -0.9567138 0.3179821gene6 -0.70287745 -1.4913486 -0.006954597 -0.6774349 1.7549044 -0.4808167gene7 0.09638077 0.5397555 1.588257406 -0.6570834 -0.3611543 0.2163203gene8 0.66215689 1.4717037 1.450286698 -0.8983272 0.4830548 -2.0012366gene9 0.20817293 1.4901270 0.598326457 -0.9547770 0.7461906 -1.0561868gene10 0.40870489 -1.8879957 0.715517402 0.2257603 0.9773011 -0.4206816> gfj2=t(gfj);gfj2gene1 gene2 gene3 gene4 gene5 gene6sample1 -0.3126880 -0.6160918 -0.4023500 0.8146320 -0.03485606 -0.702877451sample2 0.9324936 1.4331777 1.3724106 -0.2542318 -0.61820808 -1.491348558sample3 -1.1824183 -0.1901835 1.5506751 -1.3458140 -0.81190921 -0.006954597sample4 1.7039155 -0.4211536 -2.7163350 0.8897545 1.02003083 -0.677434934sample5 -0.7824128 0.6690160 0.7792040 -0.2907730 -0.95671383 1.754904412sample6 -0.2051285 0.4631609 -0.2501707 -1.0246765 0.31798207 -0.480816699gene7 gene8 gene9 gene10sample1 0.09638077 0.6621569 0.2081729 0.4087049sample2 0.53975546 1.4717037 1.4901270 -1.8879957sample3 1.58825741 1.4502867 0.5983265 0.7155174sample4 -0.65708335 -0.8983272 -0.9547770 0.2257603sample5 -0.36115431 0.4830548 0.7461906 0.9773011sample6 0.21632031 -2.0012366 -1.0561868 -0.4206816> gfj3=cbind(gfj2,rep(c('A','B'),each=3));gfj3gene1 gene2 gene3sample1 "-0.312688039051671" "-0.616091777097895" "-0.402350047076416"sample2 "0.932493636235824" "1.43317770925122" "1.37241055916329"sample3 "-1.18241825933153" "-0.190183534996257" "1.55067513562403"sample4 "1.70391552261499" "-0.421153580542317" "-2.71633500450184"sample5 "-0.782412775705917" "0.669015968383641" "0.779203971643184"sample6 "-0.205128459705795" "0.463160932158633" "-0.250170676644495"gene4 gene5 gene6sample1 "0.814631962405304" "-0.0348560593900871" "-0.702877450891742"sample2 "-0.254231813031211" "-0.61820808164723" "-1.49134855846344"sample3 "-1.34581403573095" "-0.811909209806031" "-0.00695459689994072"sample4 "0.889754527431714" "1.02003082788281" "-0.677434933681806"sample5 "-0.290772952544471" "-0.956713828084913" "1.75490441233323"sample6 "-1.02467646167772" "0.317982067115504" "-0.480816699336025"gene7 gene8 gene9sample1 "0.0963807716927326" "0.662156893062263" "0.208172932525086"sample2 "0.539755455242193" "1.47170367690911" "1.49012703771358"sample3 "1.58825740605464" "1.45028669829143" "0.598326456688562"sample4 "-0.657083351867339" "-0.898327219170378" "-0.954777049324065"sample5 "-0.361154308568598" "0.483054809616393" "0.746190587520392"sample6 "0.216320311457586" "-2.00123658452703" "-1.05618677027001"gene10sample1 "0.408704885887645" "A"sample2 "-1.88799565452592" "A"sample3 "0.715517402355723" "A"sample4 "0.225760261211124" "B"sample5 "0.977301055115831" "B"sample6 "-0.420681584016793" "B"> colnames(gfj3)[11]="group";gfj3gene1 gene2 gene3sample1 "-0.312688039051671" "-0.616091777097895" "-0.402350047076416"sample2 "0.932493636235824" "1.43317770925122" "1.37241055916329"sample3 "-1.18241825933153" "-0.190183534996257" "1.55067513562403"sample4 "1.70391552261499" "-0.421153580542317" "-2.71633500450184"sample5 "-0.782412775705917" "0.669015968383641" "0.779203971643184"sample6 "-0.205128459705795" "0.463160932158633" "-0.250170676644495"gene4 gene5 gene6sample1 "0.814631962405304" "-0.0348560593900871" "-0.702877450891742"sample2 "-0.254231813031211" "-0.61820808164723" "-1.49134855846344"sample3 "-1.34581403573095" "-0.811909209806031" "-0.00695459689994072"sample4 "0.889754527431714" "1.02003082788281" "-0.677434933681806"sample5 "-0.290772952544471" "-0.956713828084913" "1.75490441233323"sample6 "-1.02467646167772" "0.317982067115504" "-0.480816699336025"gene7 gene8 gene9sample1 "0.0963807716927326" "0.662156893062263" "0.208172932525086"sample2 "0.539755455242193" "1.47170367690911" "1.49012703771358"sample3 "1.58825740605464" "1.45028669829143" "0.598326456688562"sample4 "-0.657083351867339" "-0.898327219170378" "-0.954777049324065"sample5 "-0.361154308568598" "0.483054809616393" "0.746190587520392"sample6 "0.216320311457586" "-2.00123658452703" "-1.05618677027001"gene10 groupsample1 "0.408704885887645" "A"sample2 "-1.88799565452592" "A"sample3 "0.715517402355723" "A"sample4 "0.225760261211124" "B"sample5 "0.977301055115831" "B"sample6 "-0.420681584016793" "B"> gfj4=as.data.frame(gfj3);gfj4gene1 gene2 gene3sample1 -0.312688039051671 -0.616091777097895 -0.402350047076416sample2 0.932493636235824 1.43317770925122 1.37241055916329sample3 -1.18241825933153 -0.190183534996257 1.55067513562403sample4 1.70391552261499 -0.421153580542317 -2.71633500450184sample5 -0.782412775705917 0.669015968383641 0.779203971643184sample6 -0.205128459705795 0.463160932158633 -0.250170676644495gene4 gene5 gene6sample1 0.814631962405304 -0.0348560593900871 -0.702877450891742sample2 -0.254231813031211 -0.61820808164723 -1.49134855846344sample3 -1.34581403573095 -0.811909209806031 -0.00695459689994072sample4 0.889754527431714 1.02003082788281 -0.677434933681806sample5 -0.290772952544471 -0.956713828084913 1.75490441233323sample6 -1.02467646167772 0.317982067115504 -0.480816699336025gene7 gene8 gene9sample1 0.0963807716927326 0.662156893062263 0.208172932525086sample2 0.539755455242193 1.47170367690911 1.49012703771358sample3 1.58825740605464 1.45028669829143 0.598326456688562sample4 -0.657083351867339 -0.898327219170378 -0.954777049324065sample5 -0.361154308568598 0.483054809616393 0.746190587520392sample6 0.216320311457586 -2.00123658452703 -1.05618677027001gene10 groupsample1 0.408704885887645 Asample2 -1.88799565452592 Asample3 0.715517402355723 Asample4 0.225760261211124 Bsample5 0.977301055115831 Bsample6 -0.420681584016793 B> library(ggplot2)> zz=1:(ncol(gfj4)-1);zz[1] 1 2 3 4 5 6 7 8 9 10> gfj5=list()> for(i in zz){+ gfj5[[i]] = ggplot(data = gfj4)++ geom_boxplot(mapping = aes(x = group,+ y = colnames(gfj4)[i],+ color = group))++ geom_jitter(mapping = aes(x = group,+ y = colnames(gfj4)[i],+ color = group))++ theme_bw()+ }> library(patchwork)> wrap_plots(gfj5,ncol = 5,widths =10 ,heights = 30) #这句代码参考Sinsoledad同学
简化的代码
library(tidyverse)gfj = matrix(rnorm(60),ncol=6);gfjrownames(gfj)=paste0(rep("gene",times=10),1:10);gfjcolnames(gfj)=paste0(rep("sample",times=6),1:6);gfjgfj2 = data.frame(t(gfj))gfj3 = mutate(gfj2,group = rep(c("A","B"),each = 3))colnames(gfj3)[11]="group";gfj3class(gfj3)class(gfj3$gene1)library(ggplot2)p = list()for(i in 1:(ncol(gfj3)-1)){p[[i]] = ggplot(gfj3,aes_string(x = 'group',y = colnames(gfj3)[i]))+geom_boxplot(aes(color = group))+geom_jitter()+theme_bw()+labs(title = colnames(gfj3)[i])}library(patchwork)wrap_plots(p,ncol = 3,widths =10 ,heights = 30)
3 批量重命名(删空格)
> num=1:5> for(i in num){+ name=paste(rep("gene",times=5),i,sep=" ")+ file.create(name)+ }> filenames=list.files()> filenames[1] "20210703作业.Rproj" "20210713作业.R" "gene 1"[4] "gene 2" "gene 3" "gene 4"[7] "gene 5"> class(filenames)[1] "character"> for (i in filenames){+ newname<-sub(' ','',i)+ file.rename(i,newname)+ }
结果如下: