Genes are Discontiguous
| In “Transcribing DNA into RNA”, we mentioned that a strand of DNA is copied into a strand of RNA during transcription, but we neglected to mention how transcription is achieved. 在“将DNA转录为RNA”中,我们提到在转录过程中将一条DNA链复制到RNA链中,但是我们忽略了如何实现转录。 |
 Figure 1. The elongation of a pre-mRNA by RNAP as it moves down the template strand of DNA. |
|---|---|
| In the nucleus, an enzyme (i.e., a molecule that accelerates a chemical reaction) called RNA polymerase (RNAP) initiates transcription by breaking the bonds joining complementary bases of DNA. It then creates a molecule called precursor mRNA, or pre-mRNA, by using one of the two strands of DNA as a template strand: moving down the template strand, when RNAP encounters the next nucleotide, it adds the complementary base to the growing RNA strand, with the provision that uracil must be used in place of thymine; see Figure 1. 在细胞核中,称为RNA聚合酶(RNAP)的酶(即促进化学反应的分子)通过断开连接DNA互补碱基的键来启动转录。 然后,它使用两条DNA链中的一条作为模板链来创建一个称为前体mRNA或pre-mRNA的分子:向下移动模板链,当RNAP遇到下一个核苷酸时,它将互补碱基添加到正在生长的RNA中 ,必须使用尿嘧啶代替胸腺嘧啶; 参见图1。 |
|
| Because RNA is constructed based on complementarity, the second strand of DNA, called the coding strand, is identical to the new strand of RNA except for the replacement of thymine with uracil. See Figure 2 and recall “Transcribing DNA into RNA”. 因为RNA是基于互补性构建的,所以第二条DNA链(称为编码链)与新的RNA链相同,只是用尿嘧啶替代了胸腺嘧啶。 参见图2,并回顾“将DNA转录为RNA”。 After RNAP has created several nucleotides of RNA, the first separated complementary DNA bases then bond back together. The overall effect is very similar to a pair of zippers traversing the DNA double helix, unzipping the two strands and then quickly zipping them back together while the strand of pre-mRNA is produced. 在RNAP创建了多个RNA核苷酸后,首先分离的互补DNA碱基然后结合在一起。 总体效果非常类似于一对拉链,它们穿过DNA双螺旋,解开两条链,然后在产生前mRNA链的同时迅速将它们拉回。 |
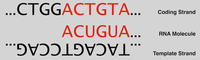 Figure 2. RNA is identical to the coding strand except for the replacement of thymine with uracil. |
| For that matter, it is not the case that an entire substring of DNA is transcribed into RNA and then translated into a peptide one codon at a time. In reality, a pre-mRNA is first chopped into smaller segments called introns and exons; for the purposes of protein translation, the introns are thrown out, and the exons are glued together sequentially to produce a final strand of mRNA. This cutting and pasting process is called splicing, and it is facilitated by a collection of RNA and proteins called a spliceosome. The fact that the spliceosome is made of RNA and proteins despite regulating the splicing of RNA to create proteins is just one manifestation of a molecular chicken-and-egg scenario that has yet to be fully resolved. 因此,并非将整个DNA子串转录成RNA,然后一次翻译成一个密码子的肽。 实际上,pre-mRNA首先被切成较小的片段,称为内含子和外显子。 为了进行蛋白质翻译,将内含子剔除,将外显子依次粘在一起以产生最终的mRNA链。 这种剪切和粘贴过程称为剪接,并且通过称为剪接体的RNA和蛋白质的集合来促进这种剪接和粘贴过程。 尽管调节RNA的剪接以产生蛋白质,但剪接体由RNA和蛋白质制成的事实只是分子鸡和蛋的情况的一种表现,该情况尚未完全解决。 In terms of DNA, the exons deriving from a gene are collectively known as the gene’s coding region. 在DNA方面,源自基因的外显子统称为基因的编码区。 |
Problem
After identifying the exons and introns of an RNA string, we only need to delete the introns and concatenate the exons to form a new string ready for translation.
Given: A DNA string s (of length at most 1 kbp) and a collection of substrings of s acting as introns. All strings are given in FASTA format.
Return: A protein string resulting from transcribing and translating the exons of s. (Note: Only one solution will exist for the dataset provided.)
Sample Dataset
Rosalind_10
ATGGTCTACATAGCTGACAAACAGCACGTAGCAATCGGTCGAATCTCGAGAGGCATATGGTCACATGATCGGTCGAGCGTGTTTCAAAGTTTGCGCCTAG
>Rosalind_12
ATCGGTCGAA
>Rosalind_15
ATCGGTCGAGCGTGTSample Output
Solution
本题讲的是真核生物的 RNA 选择性剪切过程,题目要求输入多个内含子(introns),将该 DNA 序列的内含子(introns)全部切除后的外显子(exons)构成一段编码 RNA;然后求出这段编码 RNA 翻译得到的蛋白质序列。
将 DNA 上内含子全部删除,ATGGTCTACATAGCTGACAAACAGCACGTAGCA|--------|TCTCGAGAGGCATATGGTCACATG|-------------|TTCAAAGTTTGCGCCTAGATCGGTCGAA ATCGGTCGAGCGTGT后链接 3 段外显子ATGGTCTACATAGCTGACAAACAGCACGTAGCA TCTCGAGAGGCATATGGTCACATG TTCAAAGTTTGCGCCTAG <--/转录AUGGUCUACAUAGCUGACAAACAGCACGUAGCAUCUCGAGAGGCAUAUGGUCACAUGUUCAAAGUUUGCGCCUAG||| 翻译 ***---$$$---$$$---$$$---$$$---$$$---$$$---$$$---$$$---$$$---$$$---$$$---算法流程:
- 对所有内含子进行剪切操作,得到
x段外显子。 - 将
x段外显子连接,进行转录+翻译。
我们可以使用正则表达式进行一次性 split 操作吗,然后拼接后转录翻译即可。
from typing import Listimport reclass Solution:def RNASplicing(self, DNA: str, introns: List[str], codon: dict) -> str:exons = ''.join(re.split('|'.join(introns), DNA))RNA = exons.replace('T', 'U') # 翻译(因为 exons 是编码链)protein = []for start in range(RNA.find('AUG'), len(RNA), 3):if codon[RNA[start:start+3]] != 'Stop':protein.append(codon[RNA[start:start+3]])else: breakreturn ''.join(protein)codonTable = """UUU F CUU L AUU I GUU VUUC F CUC L AUC I GUC VUUA L CUA L AUA I GUA VUUG L CUG L AUG M GUG VUCU S CCU P ACU T GCU AUCC S CCC P ACC T GCC AUCA S CCA P ACA T GCA AUCG S CCG P ACG T GCG AUAU Y CAU H AAU N GAU DUAC Y CAC H AAC N GAC DUAA Stop CAA Q AAA K GAA EUAG Stop CAG Q AAG K GAG EUGU C CGU R AGU S GGU GUGC C CGC R AGC S GGC GUGA Stop CGA R AGA R GGA GUGG W CGG R AGG R GGG G"""# 1. 创建字符频率表codon = {}for line in codonTable.splitlines():line = line.split()for i in range(0, len(line), 2):codon[line[i]] = line[i+1]fasta = """>Rosalind_10ATGGTCTACATAGCTGACAAACAGCACGTAGCAATCGGTCGAATCTCGAGAGGCATATGGTCACATGATCGGTCGAGCGTGTTTCAAAGTTTGCGCCTAG>Rosalind_12ATCGGTCGAA>Rosalind_15ATCGGTCGAGCGTGT"""DNA, *introns = (seq.replace('\n', '') for seq in re.split(r'>.*', fasta) if seq.replace('\n', '')) # 第一个是 `\n`print(Solution().RNASplicing(DNA, introns, codon))

