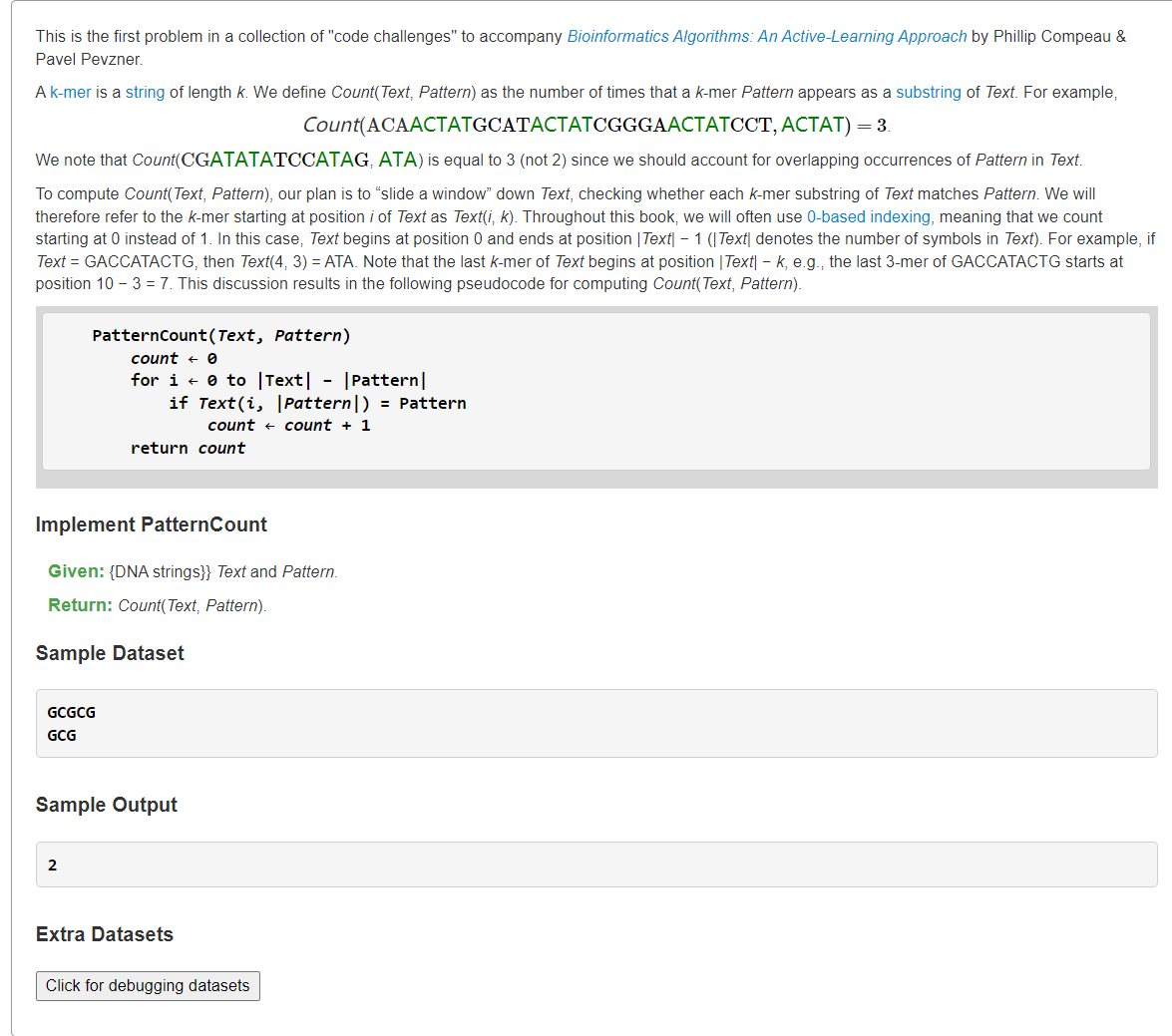
- Compute the Number of Times a Pattern Appears in a Textoverlap">Compute the Number of Times a Pattern Appears in a Textoverlap
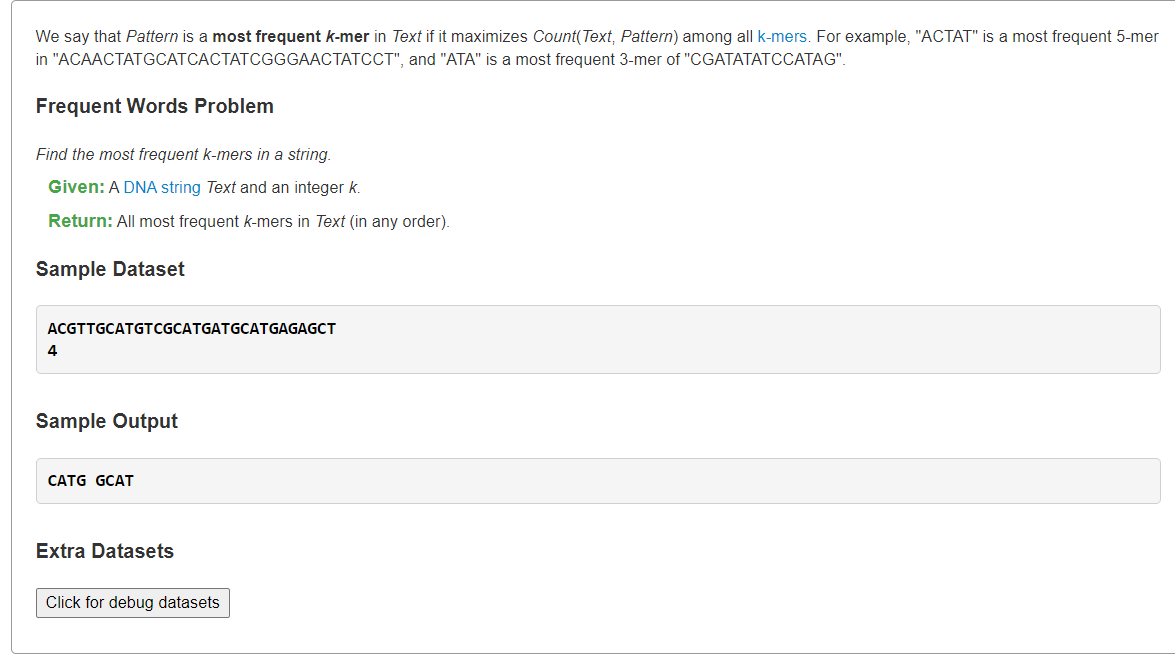
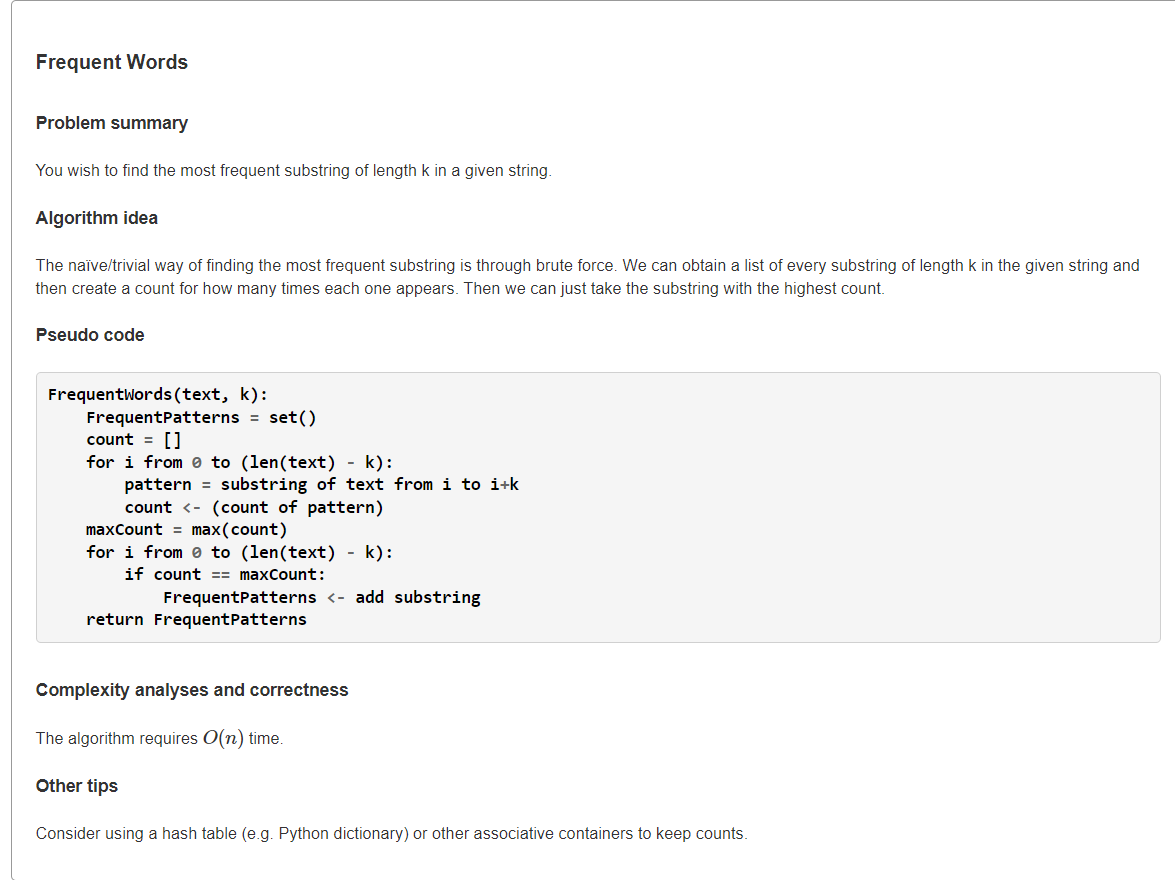
- Find the Most Frequent Words in a Stringoverlap">Find the Most Frequent Words in a Stringoverlap
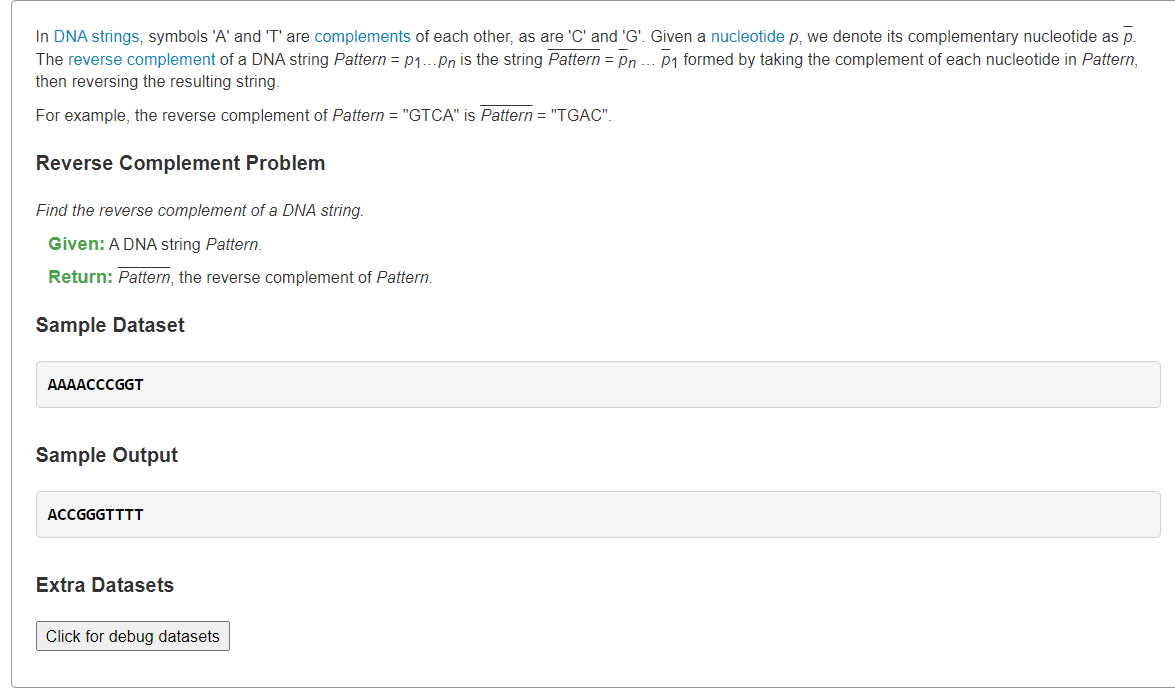
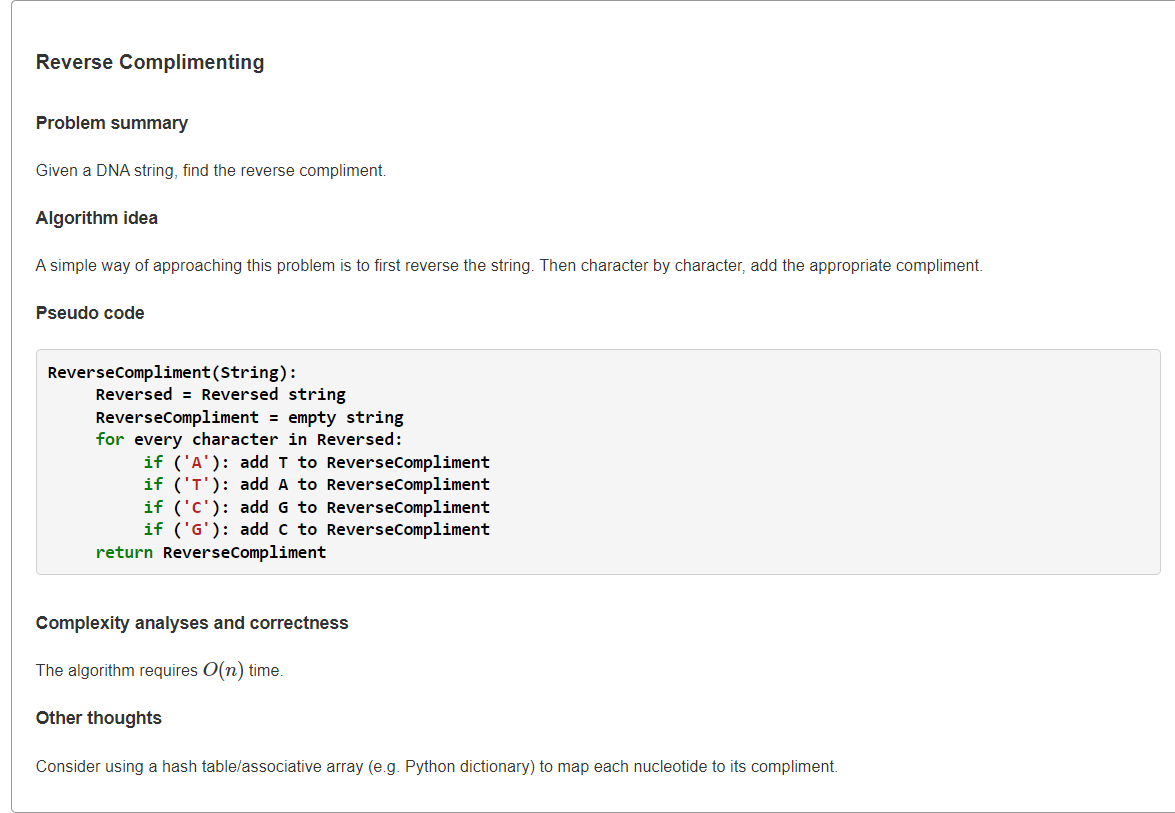
- Find the Reverse Complement of a String">Find the Reverse Complement of a String
- Find All Occurrences of a Pattern in a Stringoverlap">Find All Occurrences of a Pattern in a Stringoverlap
- Find Patterns Forming Clumps in a String">Find Patterns Forming Clumps in a String
- Find a Position in a Genome Minimizing the Skew">Find a Position in a Genome Minimizing the Skew
- Find All Approximate Occurrences of a Pattern in a String">Find All Approximate Occurrences of a Pattern in a String
- Generate the d-Neighborhood of a StringBFS/DFS">Generate the d-Neighborhood of a StringBFS/DFS
- Implement PatternToNumber字符串转对应四进制数">Implement PatternToNumber字符串转对应四进制数
- Implement NumberToPattern四进制数转对应字符串">Implement NumberToPattern四进制数转对应字符串
- Find the Most Frequent Words with Mismatches in a String4^k个k-mer频数表">Find the Most Frequent Words with Mismatches in a String4^k个k-mer频数表
- Find the Most Frequent Words with Mismatches in a String4^k个k-mer汉明距离≤d">Find the Most Frequent Words with Mismatches in a String4^k个k-mer汉明距离≤d
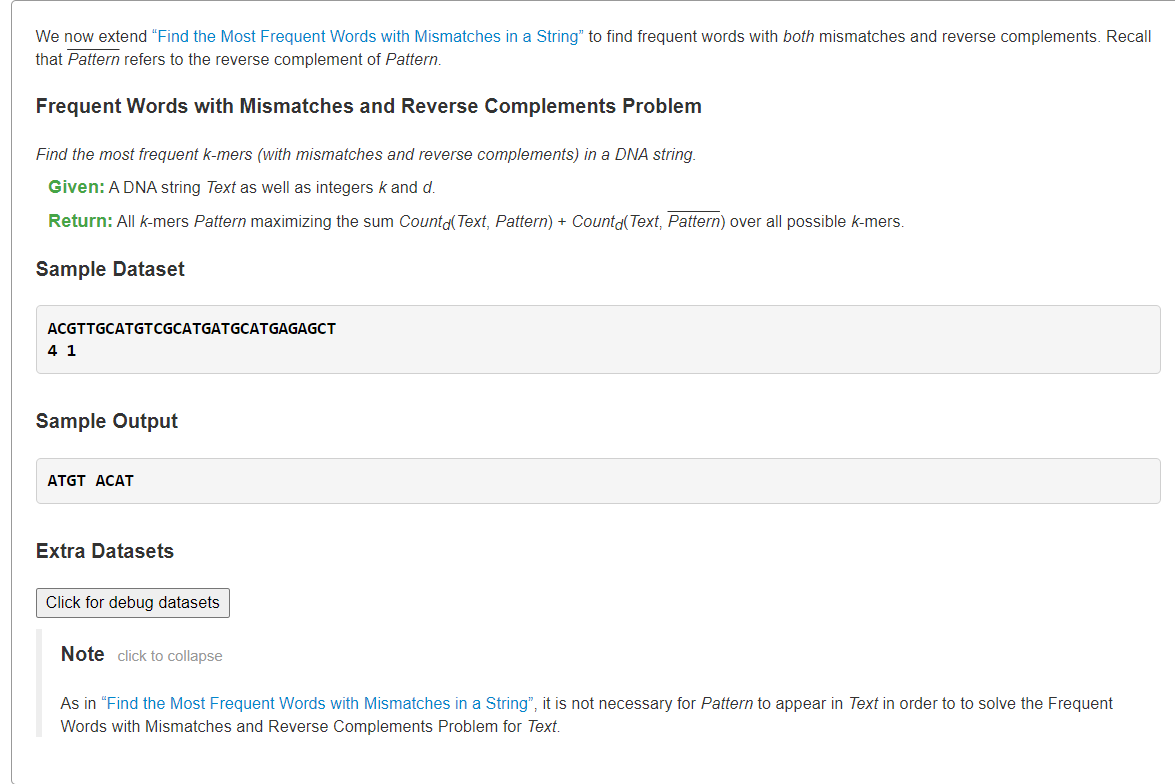
- Find Frequent Words with Mismatches and Reverse Complements">Find Frequent Words with Mismatches and Reverse Complements
Compute the Number of Times a Pattern Appears in a Textoverlap
1、暴力:O(|t||p|)
如题干的算法伪代码所示:
def patternCount(text: str, pattern: str) -> int:"""Count overlapping substring in a given string"""m, n = len(text), len(pattern)count = 0for i in range(0, m - n + 1):if text[i:i+n] == pattern:count += 1return counts = """GCGCGGCG"""print(patternCount(*s.splitlines()))
pub fn count_pattern(text: &str, pattern: &str) -> u32 {let mut count = 0;let num = pattern.len();let length = text.len();// println!("Length: {}/{}", length, num);for (idx, _) in text.chars().enumerate() {let max_len = idx + num;if max_len < length {match &text[idx..(idx + num)] == pattern {true => count = count + 1,_ => continue,}}}return count;}
2、strstr
public class KMP {public static void main(String[] args) {int[] ababas = kmp ("ab");System.out.println (Arrays.toString (ababas));int i = kmpSearch ("abababababa", "aba", ababas);System.out.println (i);}public static int[] kmp(String s){ //AAADAAAADDAint[] temp = new int[s.length ()];temp[0] = 0;for (int i = 0,j=1; j < s.length (); j++) {while (i>0&&s.charAt (i)!=s.charAt (j)){i = temp[i-1];}if (s.charAt (i)==s.charAt (j)) {i++;}temp[j] = i;}return temp;}public static int kmpSearch(String str1,String str2,int[] next){//遍历str1int ans = 0;for (int i = 0,j = 0; i < str1.length (); i++) {while (j>0&&str1.charAt (i)!=str2.charAt (j)){j = next[j-1];}if (str1.charAt (i)==str2.charAt (j)){j++;}if (j==str2.length ()&&j>=2){ans++;j = next[j-2];i--;}}return ans;}}作者:Condescending ColdenjK1链接:https://leetcode.cn/circle/discuss/Cqoohu/view/FHApDf/来源:力扣(LeetCode)著作权归作者所有。商业转载请联系作者获得授权,非商业转载请注明出处。
3、SA+Height
后缀数组+height数组,将t#s连起来后得后缀数组,排序获得height,然后找s出现位置。
Find the Most Frequent Words in a Stringoverlap
1、暴力:O(|t|2|k|)
枚举中所有长度为
的子串作为
,然后调用
计算该串重叠出现次数。注意:由于
中所有长度为
的子串可能有重复,因此若它们出现次数都是最大,最终输出要去重。实现上就是用集合记录最频繁出现的
from typing import Listdef patternCount(text: str, pattern: str) -> int:"""Count overlapping substring in a given string"""m, n = len(text), len(pattern)count = 0for i in range(0, m - n + 1):if text[i:i+n] == pattern:count += 1return countdef mostFrequentWord(text: str, k: int) -> List[str]:"""Find the Most Frequent Words in a String"""ans, cnt = {}, 0 # pattern may be repeat, so we use set to deduplicate.for i in range(len(text) - k + 1):# enumerate all k-mer substringpattern = text[i:i+k]count = patternCount(text, pattern)if cnt < count:ans = {pattern}cnt = countelif cnt == count:ans.add(pattern)return list(ans)s = """ACGTTGCATGTCGCATGATGCATGAGAGCT4"""text, k = s.splitlines()print(*mostFrequentWord(text, int(k)))
dna <- ("ACGTTGCATGTCGCATGATGCATGAGAGCT")n <- 4kmer.list <- numeric(nchar(dna)-(n-1))for (i in 1:(nchar(dna)-(n-1))){kmer.list[i] <- substring(dna, i, (i+(n-1)))}kmer.list <- as.matrix(subset(kmer.list, kmer.list > 0))kmer.list <- as.matrix(sort(table(kmer.list), decreasing = TRUE))frequent_words <- function(text, k) {frequentpatterns <- matrix()textunlisted <- unlist(strsplit(text, split=""))n <- nchar(text)for (i in 1:(n-k+1)) {pattern <- paste(textunlisted[i:(i+k-1)], collapse="")count[[i]] <- matrix(pattern_count(text,pattern))}maxcount <- max(count)for (i in 1:(n-k+1)){if(count[[i]] == maxcount)frequentpatterns[[i]] <- matrix(paste(textunlisted[i:(i+k-1)], collapse=""))}frequentpatterns <- unique(frequentpatterns)frequentpatterns <- frequentpatterns[!is.na(frequentpatterns)]return(frequentpatterns)}
2、k长度滑窗+滚动哈希+计数器:O(|t|+|k|)
from typing import Listfrom collections import Counterdef mostFrequentWord(text: str, k: int) -> List[str]:"""Find the Most Frequent Words in a String"""cnt = Counter(text[i:i+k] for i in range(len(text) - k + 1))maxCnt = max(cnt.values())return [s for s, v in cnt.items() if v == maxCnt]s = """ACGTTGCATGTCGCATGATGCATGAGAGCT4"""text, k = s.splitlines()print(*mostFrequentWord(text, int(k)))
哈希表可以通过 排序+双指针 实现计数
3、SA
Explanation
Find the Reverse Complement of a String
1、模拟
def reverseComplementOfDNA(dna: str) -> str:"""Find the Reverse Complement of a String"""# return dna.translate(str.maketrans('AGCT', 'TCGA'))[::-1]d = dict(zip('AGCT', 'TCGA'))n = len(dna)ret = [''] * nfor i in range(n):ret[n-1-i] = d[dna[i]]return "".join(ret)print(reverseComplementOfDNA("AAAACCCGGT"))
Explanation
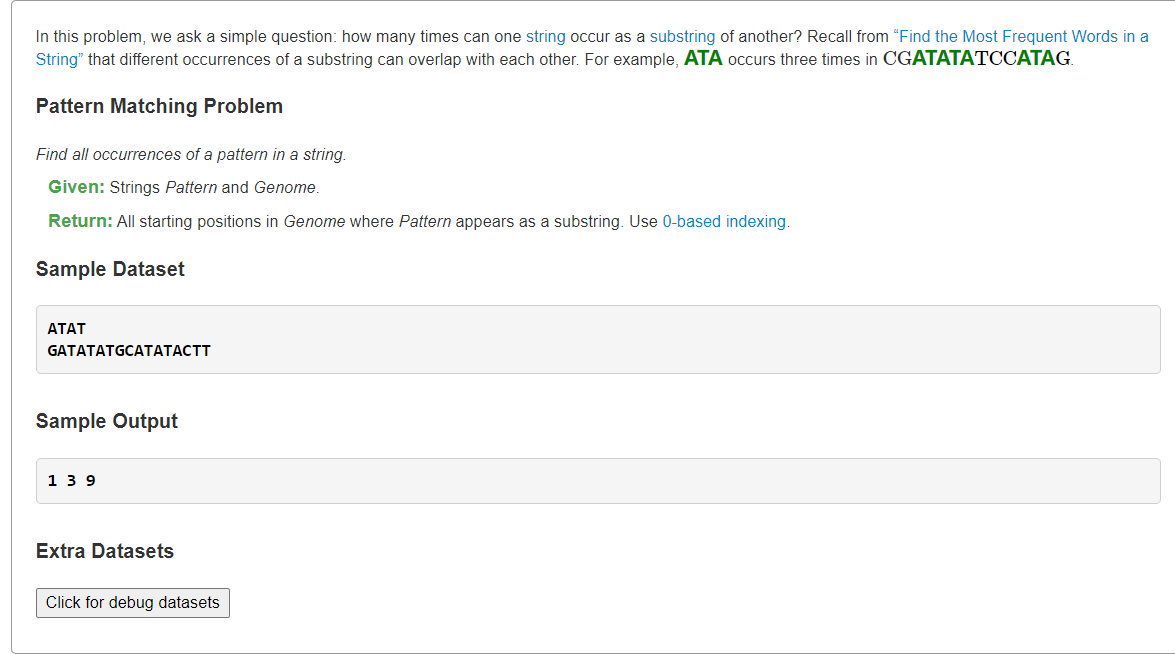
Find All Occurrences of a Pattern in a Stringoverlap
1、KMP找出所有匹配位置
from typing import Listdef strstr(t: str, p: str) -> List[int]:"""Find all the occurence place of p in t"""m, n = len(t), len(p)if n == 0:return [0]# calculate next arraynext = [0] * ni, j = 1, 0while i < n:while j > 0 and p[i] != p[j]:j = next[j-1]if p[i] == p[j]:j += 1next[i] = ji += 1# print(next)# KMPidxs = []i, j = 0, 0while i < m:while j > 0 and t[i] != p[j]:j = next[j-1]if t[i] == p[j]:j += 1if j == n:# we can return i - n + 1 if find the first occurence place,# otherwise assume t[i] != p[n-1]idxs.append(i - n + 1)j = next[n-1]i += 1# print(idxs)return idxss = """ATATGATATATGCATATACTT"""p, t = s.splitlines()print(*strstr(t, p))
fn main() {let pattern = "ATAT";let genome = String::from("GATATATGCATATACTT");let positions = find_pattern_positions(pattern, genome) ;for pos in positions {print!("{} ",pos)}}fn find_pattern_positions(pattern: &str, genome:String) -> Vec<u32> {let mut positions: Vec<u32> = Vec::new();for i in 0..genome.len()-pattern.len(){if genome[i..i+pattern.len()] == *pattern {positions.push(i as u32);}}positions}
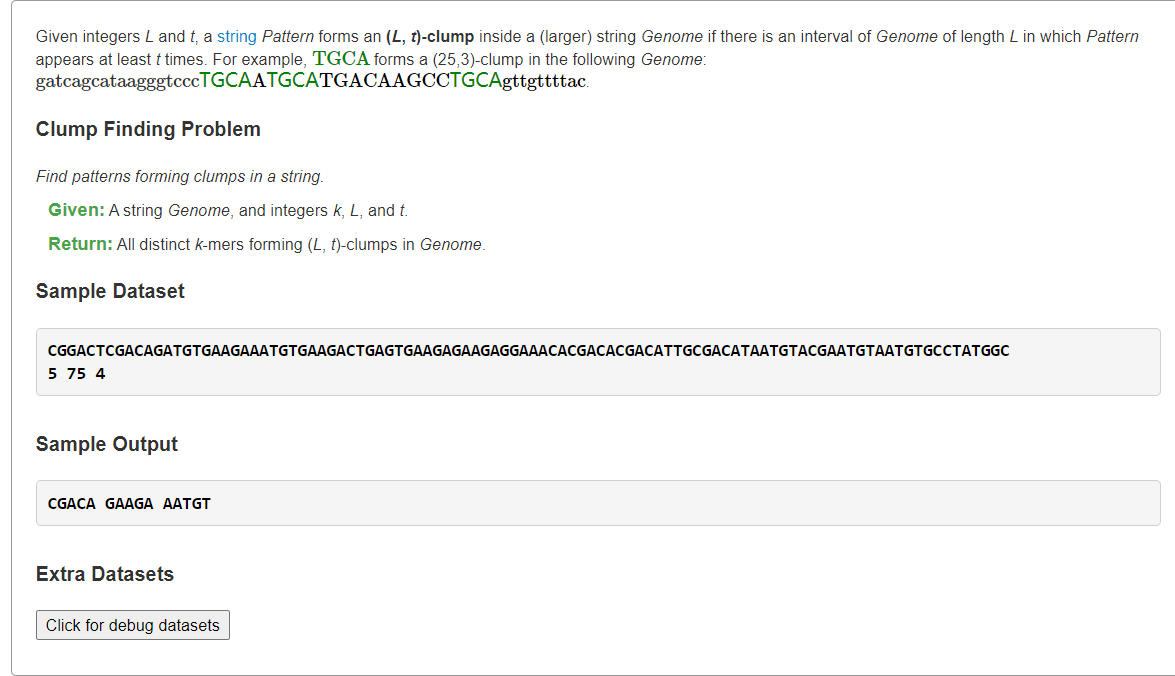
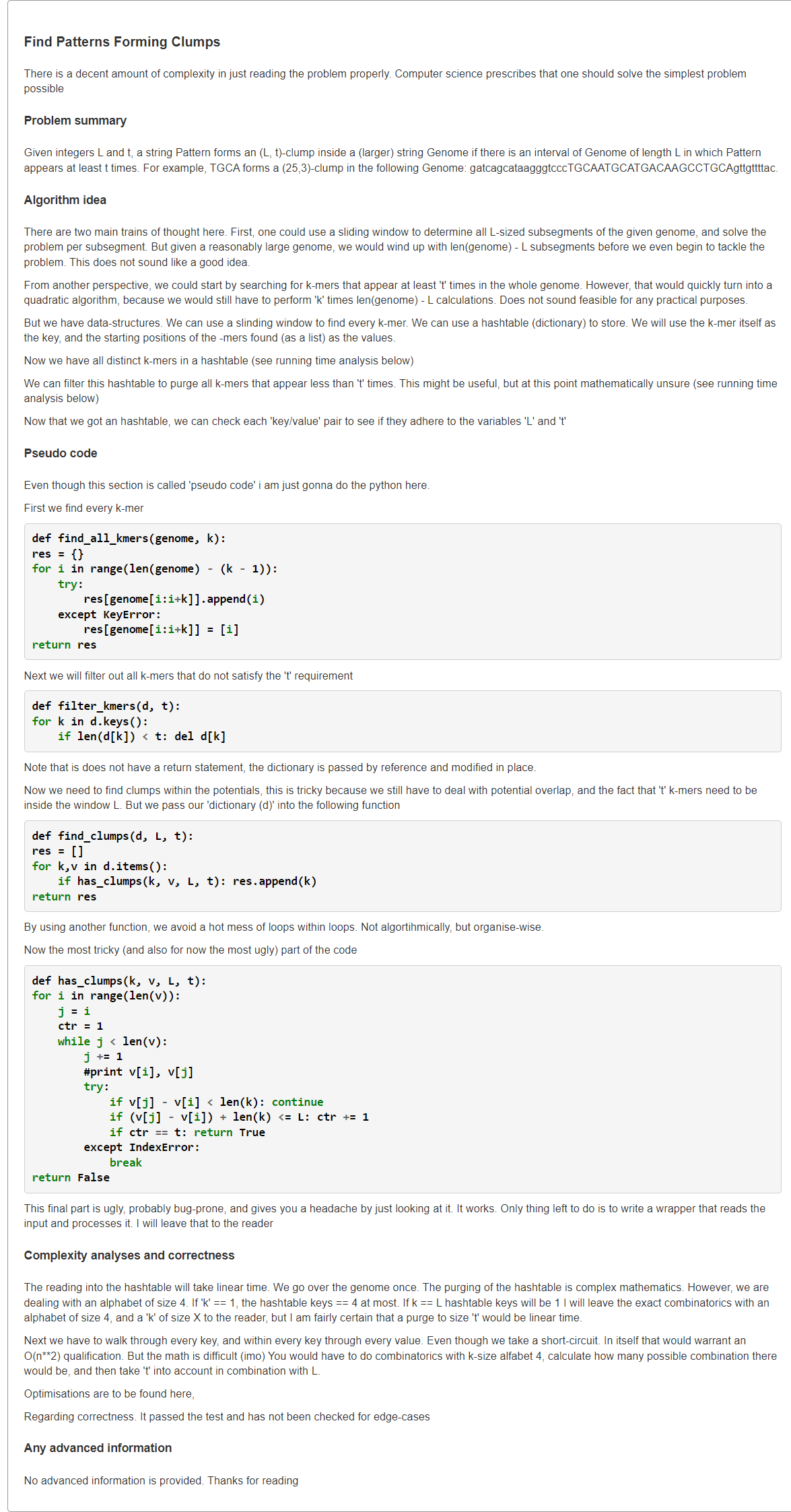
Find Patterns Forming Clumps in a String
1、暴力:O(L2k|Genome|)
def FrequencyTable(text, k):freq_map = {}n = len(text)for i in range(0, n - k + 1):pattern = text[i:i + k]if pattern not in freq_map:freq_map[pattern] = 1else:freq_map[pattern] = freq_map[pattern] + 1return freq_mapdef FindClumps(text, k, l, t):patterns = set()n = len(text)for i in range(n - l + 1):window = text[i:i + l]freq_map = FrequencyTable(window, k)for s in freq_map:if freq_map[s] >= t:patterns.add(s)return ' '.join(patterns)filename = 'rosalind_ba1e.txt'file = open(filename, 'r')lines = file.readlines()text = lines[0].strip()params = lines[1].strip().split()k, l, t = [int(p) for p in params]print(FindClumps(text, k, l, t))
2、所有k-mer出现位置+第i个出现位置到第i+t个出现位置的距离是否≤L
from typing import Listdef clumpFind(genome: str, k: int, L: int, t: int) -> List[str]:# O(nk): enumerate k-mer in genomen = len(genome)kmerIdxs = {} # {k-mer: indexs}for i in range(n - k + 1):p = genome[i:i+k] # can be optimize with rolling hash# add the occurrence of p in GenomekmerIdxs.setdefault(p, []).append(i)ans = []# 时间复杂度较难计算,但<<O(n^2)# 对每个k-mer出现位置,看L长度内有多少个(滑窗O(|idx|)解决)# if the number overlap occurence idx >= tfor s, idx in kmerIdxs.items():left = 0for right in range(len(idx)):# slide in idx[right]# check the length of idx[left]~idx[right]while left <= right and idx[right] - idx[left] + 1 > L:left += 1if right - left + 1 >= t:ans.append(s)breakreturn anss = """CGGACTCGACAGATGTGAAGAAATGTGAAGACTGAGTGAAGAGAAGAGGAAACACGACACGACATTGCGACATAATGTACGAATGTAATGTGCCTATGGC5 75 4"""lines = s.splitlines()genome = lines[0]k, L, t = map(int, lines[1].split())print(*clumpFind(genome, k, L, t))
3、评论区最优解法:长度为L的滑窗(初始化+滑入/出一个k-mer)
本题可以overlap,左侧滑出,那么起点为
的 k-mer 数少一,滑入
,那么起点为
的 k-mer 数加一,窗口内的所有起点
的 k-mer 数不变,因此每次滑动只需要判断新进来的起点为
的 k-mer 数是否 ≥ t
总时间复杂度为
- 初始化滑窗:枚举长度为
窗口内所有长度为
的子串需要
,可用滚动哈希优化为
- 每次滑动转移需要
计算滑入/出的k-mer串的哈希值,时间复杂度为
:::info 进阶:非overlap
30. 串联所有单词的子串,时间复杂度为O(k×|G|k) :::Explanation
Find a Position in a Genome Minimizing the Skew
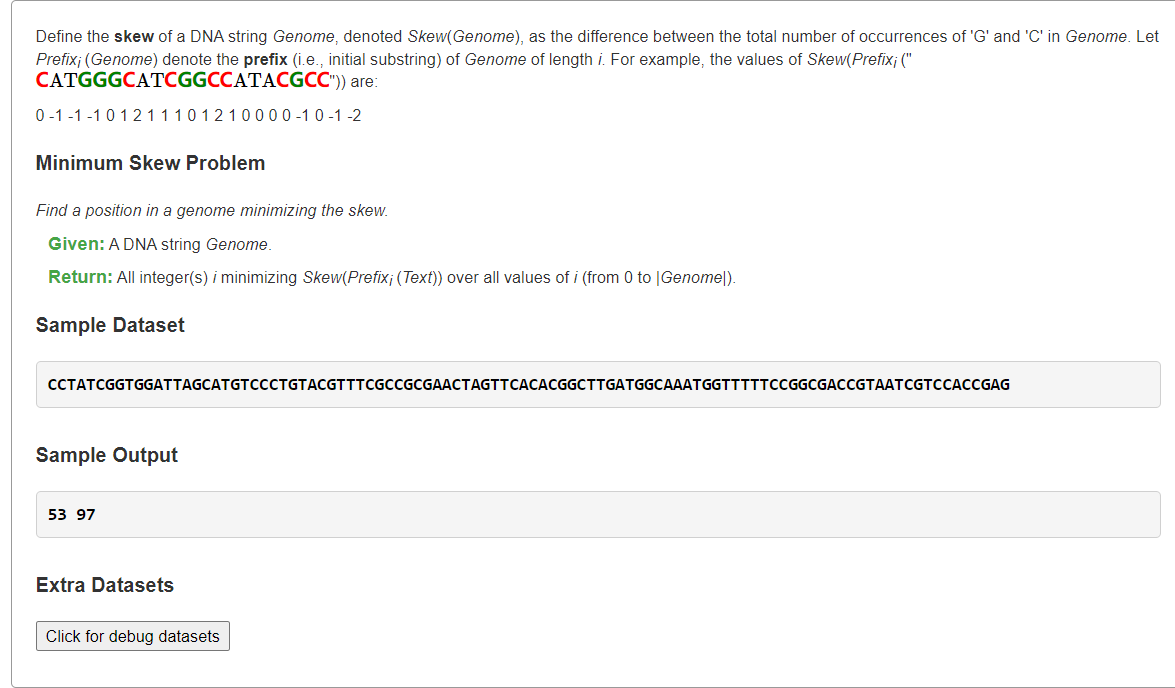
1、G,C目前累计数量->G-C差值
```python from typing import List
def minimumSkew(dna: str) -> List[int]: n = len(dna) minv, ans = n, [] G = C = 0 for i in range(n): if dna[i] == ‘G’: G += 1 elif dna[i] == ‘C’: C += 1 if G - C < minv: ans = [i + 1] # next place is oriC minv = G - C elif G - C == minv: ans.append(i + 1) # next place is oriC return ans
s = “””CCTATCGGTGGATTAGCATGTCCCTGTACGTTTCGCCGCGAACTAGTTCACACGGCTTGATGGCAAATGGTTTTTCCGGCGACCGTAATCGTCCACCGAG”””
print(*minimumSkew(s))
<a name="CkIoo"></a># [Compute the Hamming Distance Between Two Strings](https://rosalind.info/problems/ba1g/)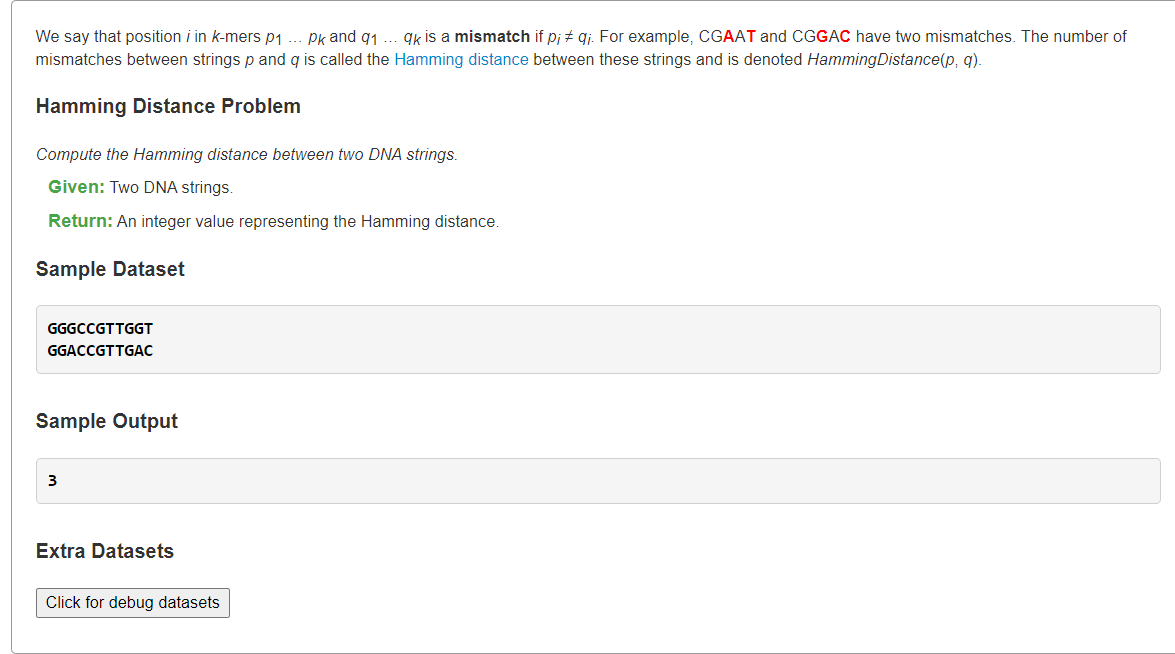<a name="YcT8P"></a>## 1、汉明距离:统计每位s[i]!=t[i]总数```pythondef hammingDistance(s: str, t: str) -> int:dist = 0for ch1, ch2 in zip(s, t):if ch1 != ch2:dist += 1return dists = """GGGCCGTTGGTGGACCGTTGAC"""s, t = s.splitlines()print(hammingDistance(s, t))
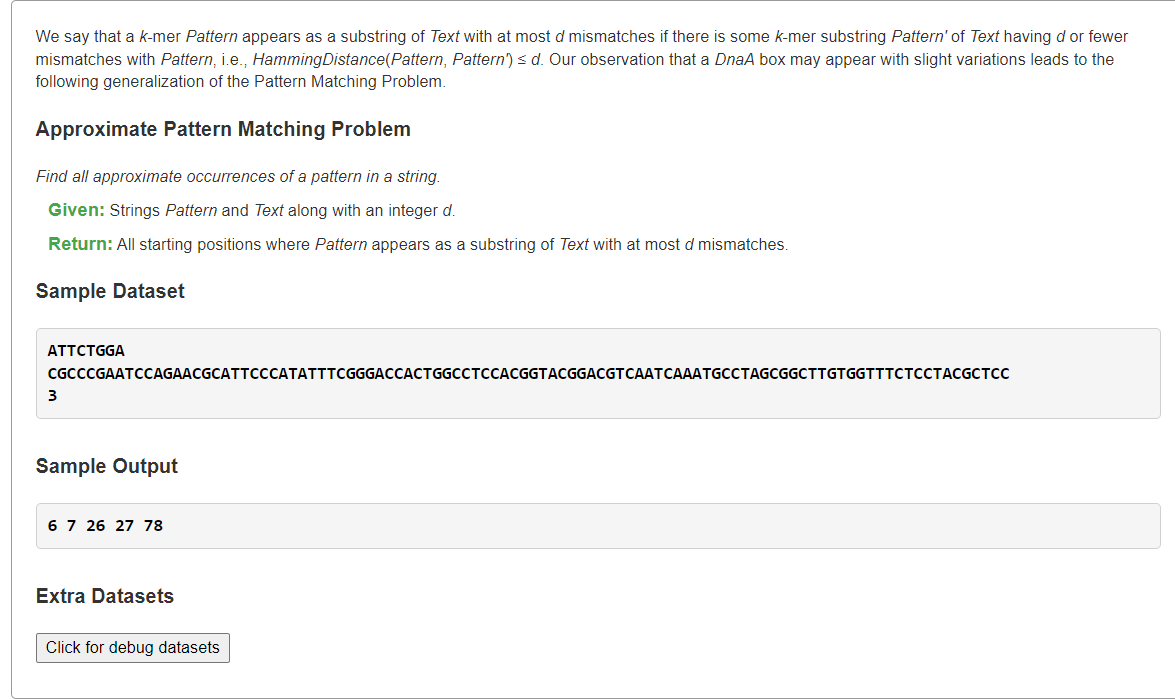
Find All Approximate Occurrences of a Pattern in a String
1、暴力解:O(|T||P|)
枚举text中所有长度为的子串
, 检验其与
的汉明距离是否小于等于d,是就将索引
加入答案。
from typing import Listdef hammingDistance(s: str, t: str) -> int:dist = 0for ch1, ch2 in zip(s, t):if ch1 != ch2:dist += 1return distdef approximatePatternMatch(text: str, pattern: str, d: int) -> List[int]:m, n = len(text), len(pattern)ans = []for i in range(m - n + 1):if hammingDistance(text[i:i+n], pattern) <= d:ans.append(i)return anss = """ATTCTGGACGCCCGAATCCAGAACGCATTCCCATATTTCGGGACCACTGGCCTCCACGGTACGGACGTCAATCAAATGCCTAGCGGCTTGTGGTTTCTCCTACGCTCC3"""pattern, text, d = s.splitlines()print(*approximatePatternMatch(text, pattern, int(d)))
2、待更新DP
允许错配
https://en.wikipedia.org/wiki/Approximate_string_matching
https://www.cs.unc.edu/~prins/Classes/555/Media/Lec18.pdf
测试用例:
https://bioinformaticsalgorithms.com/data/debugdatasets/replication/ApproximatePatternMatchingProblem.pdf
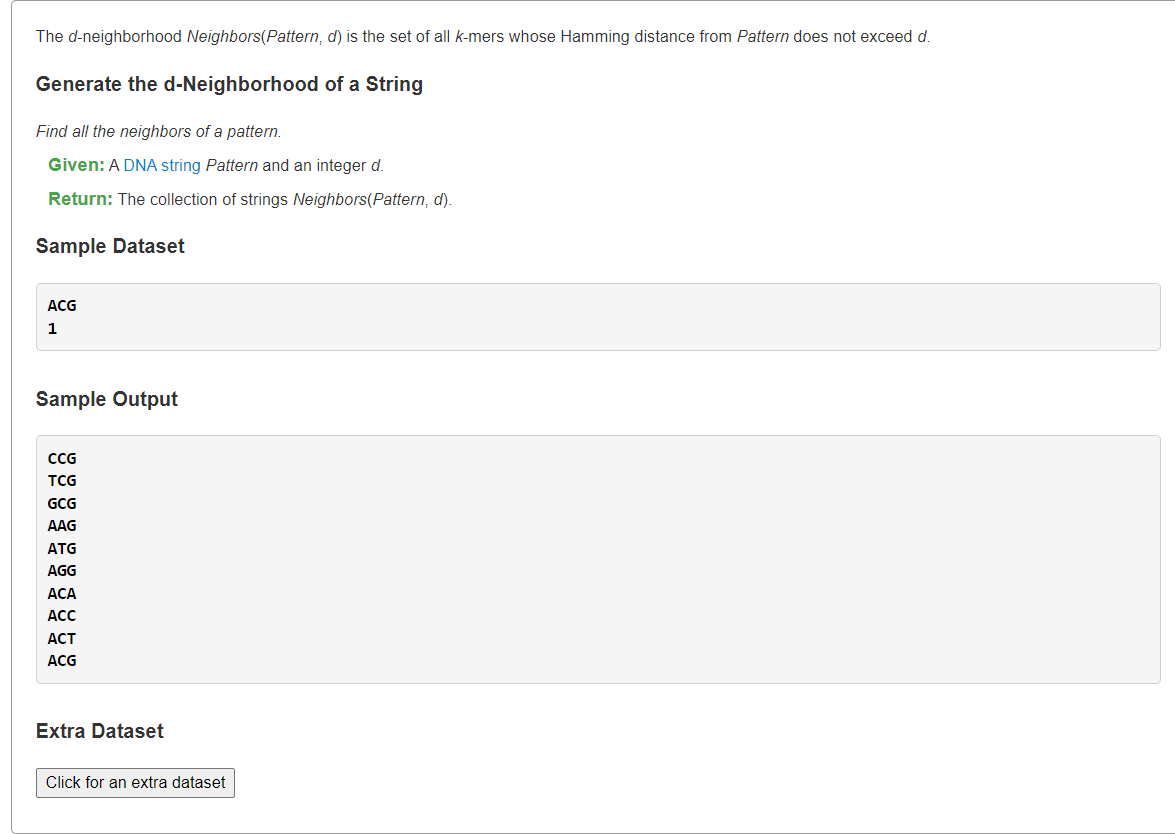
Generate the d-Neighborhood of a StringBFS/DFS
1、枚举所有长度为|Pattern|的组成可能+汉明距离筛选
最直观的方式就是生成个所有可能序列,然后在它们中筛出与
的汉明距离≤d的构成答案。
def get_d_neighbors(pattern: str, d: int) -> str:return "\n".join([xfor x in ("".join(i)for i in itertools.product(["A", "T", "G", "C"], repeat=len(pattern)))if hamming_distance(pattern, x) <= d])
复杂度分析:
- 时间复杂度:
,一次汉明距离筛选需要
,总共进行
次
-
2、记录枚举树中所有节点
:::info 书本上代码允许一个位置
突变多次,且允许
突变为自身,因此可能出现
s[i]->...->s[i],导致产生很多重复集,最终结果需要使用集合去重。
如果我们限制最多突变一次且只能突变为非
字符,就能保证每次突变产生不同结果。 ::: 如下图
s = ACG, d = 2的枚举树,层序号就是与s的汉明距离,例如第1层表示其与s距离为1。其中淡蓝色的节点是重复出现,去重方式就是保证突变位置递增,因此所求答案就是所有非重复树节点。
复杂度分析:设长度为
,字符集
,汉明距离差为
时间复杂度:
,分别枚举突变次数从
,第
次突变可发生在
个位置之一,该次突变结果有
种(不允许变成自己),第
次突变可发生在
个位置之一(不允许一个位置突变多次),该次突变结果有
种(不允许变成自己),因此突变数为
时,产生
种结果。
- 空间复杂度:
实现1:DFS/回溯
评论区的DFS算法,其去重方式要求每次修改位置必须是递增,因此其枚举树就是上图 ```python def Neighbors(Pattern, k, d, start = 0): neighbors = [Pattern] # 初始化把自己加入 if d > 0:
return neighborsfor i in range(start, k):for symbol in "ACGT":if symbol != Pattern[i]:new_pattern = Pattern[:i] + symbol + Pattern[i+1:]new_neighbors = Neighbors(new_pattern, k, d - 1, i + 1) # 注意i+1neighbors.extend(new_neighbors)
kmer = input() d = int(input()) neighbors = Neighbors(kmer, len(kmer), d) for neighbor in neighbors: print(neighbor)
01选取:对于每个位置有两种选择,突变或不突变:- 不突变:那么总突变数不变,继续向后求解中总突变数为的结果- 突变:前提是,当前突变结果有三种(下面代码规定只允许一次有效突变:不能突变两次或突变一次变回),每种使用回溯选择一下,继续向后求解中总突变数为的结果```pythonfrom typing import Listdef neighbors(pattern: str, d: int) -> List[str]:s = list(pattern) # 转数组可修改ans = [] # 回溯生成不重复,每个s[i]只能变为字符集内其它字符def backtrace(idx: int, d: int) -> None: # 去重一:保证idx递增# 递归出口if idx >= len(s) or d == 0:ans.append("".join(s))return# 当前选择不变backtrace(idx + 1, d)# 当前选择突变if d > 0:orginBase = s[idx]for base in "ACGT":if base != orginBase: # 去重二:保证s[i]变为其它字符s[idx] = base # 选择backtrace(idx + 1, d - 1)s[idx] = orginBase # 撤销backtrace(0, d)# print(len(ans) == len(set(ans)))return anss = """ACG1"""s, d = s.splitlines()print(*neighbors(s, int(d)), sep='\n')
实现2:BFS【从src向下搜d层】
从上往下一层层搜索,但是怎么去重呢?
def mutation(s, d):ans = set()queue = []dist = {}queue.append(s)# 此dist字典没必要,其就是记录BFS层数,使用layer变量记录即可dist[s] = 0 # d[t]表示s->...->t共花费几步(允许同位置变多次+变为自己)ans.add(s) # 使用集合去重while len(queue) != 0:cur = queue.pop(0) # 可以改为collections.dequefor i in range(len(cur)):cur_l = list(cur)for j in bp: # 允许s[i]→s[i],因此会产生重复解cur_l[i] = jnew_str = "".join(cur_l)#print(new_str)# 使用ans标记重复,防止重复节点搜索,也就是s[i]只能变为非s[i]字符if new_str not in ans and dist[cur] + 1 <= d:ans.add(new_str)queue.append(new_str)dist[new_str] = dist[cur] + 1return ans
上面代码使用来标记访问过的节点,这样可以保证不重复,即最终所求答案,但是
dist字典根本没用
我们可以使用每层突变位置必须是递增的方式来去重,也就是节点设计为(src, mutPos),下一个节点突变位置只能在mutPos + 1 -> |src|-1。
from typing import Listdef neighbors(pattern: str, d: int) -> List[str]:ans = [pattern] # d=0将自己加入from collections import dequeq = deque([(pattern, -1)])while d:for _ in range(len(q)):u, mutPos = q.popleft()# 突变位置递增且s[i]只能变为字符集内其它字符可保证生成结果不重复for i in range(mutPos + 1, len(u)):for base in "ACGT":if u[i] != base:v = u[:i] + base + u[i+1:]ans.append(v) # 记录入队节点q.append((v, i))# print("next layer: ", q)d -= 1# print(len(ans) == len(set(ans)))return anss = """ACG2"""s, d = s.splitlines()print(*neighbors(s, int(d)), sep='\n')
Explanation
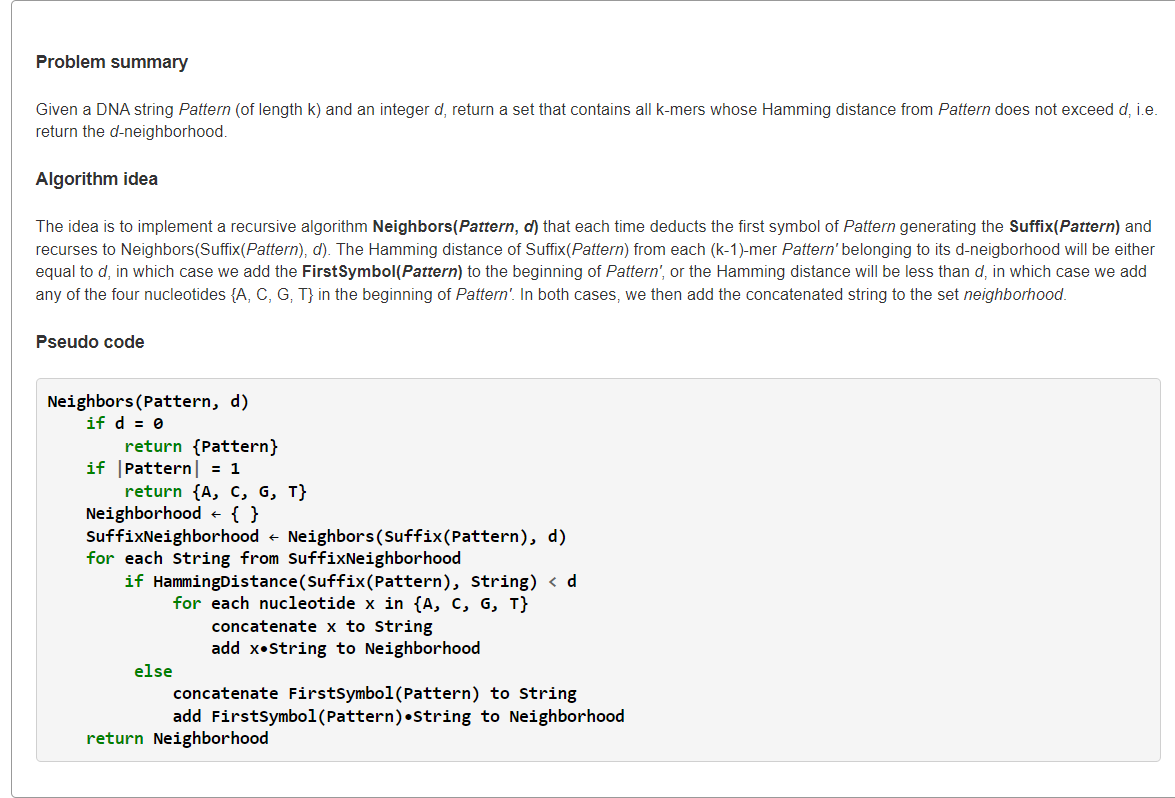
书本上的代码是允许多次突变+允许
突变回
,这样就会出现大量重复解,因此需要用集合去重。
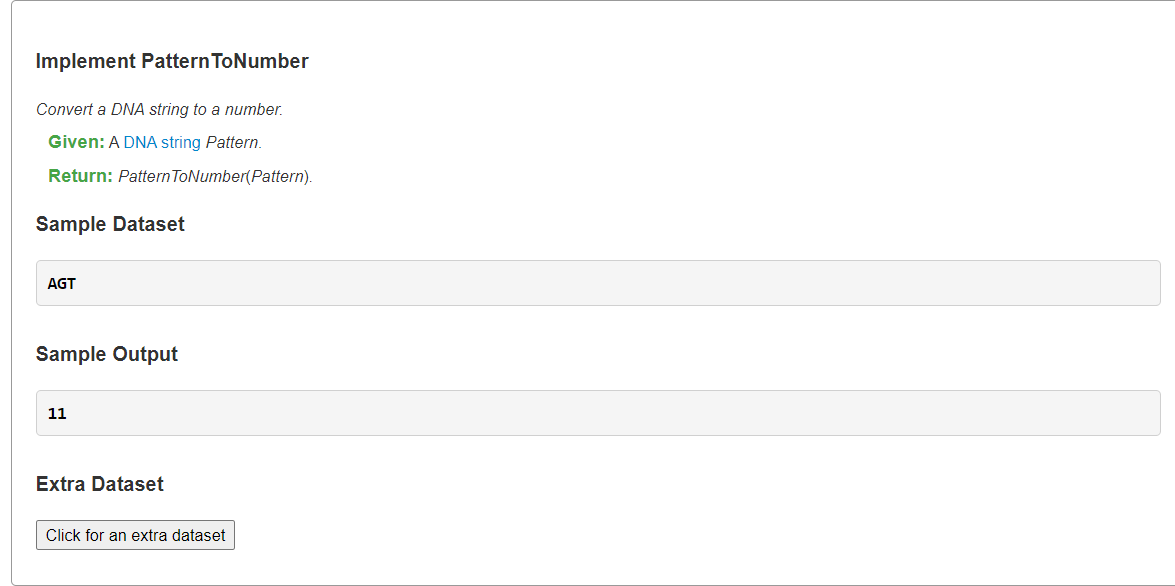
Implement PatternToNumber字符串转对应四进制数
1、分支界定法:x是字典序第几(从0开始计数)
AGT排第几?前缀AG位于第二层且序号为2,即左侧有2个兄弟节点,它们每个可向下生长N=4个分支,因此AG的第一个孩子序号为2 * N,而T是AG的第4个字典序孩子,因此其序号为2 * N + 3
AGTG排第几?前缀AGT位于第三层且序号在为11,即左侧有11个兄弟节点,它们每个可向下生长N=4个分支,因此共AGT的的第一个孩子序号为11 * N,而G是AGT的第3个字典序孩子,因此其序号为11 * N + 2
综上有公式:
当前串的序号 = prefix的序号 分支个数N + 当前字符ch的字典序
那么prefix串的序号怎么求呢?其可以继续带入上面公式,因此这个是递归问题。
递归函数定义:返回
串在由字符集
构成的字典序顺序(从0开始计数)
递归出口:当时,直接看该字符在
排第几返回;实际也可将
时返回
当作出口,因为其不影响结果。
递归链条:若,拆分除前缀
和末尾字符
,当前
的序号 =
序号 分支个数N + 当前字符
的字典序
symbol2index = dict(zip('ACGT', range(4)))def pattern2number(pattern: str) -> int:if not pattern:return 0number = symbol2index[pattern[-1]]return 4 * pattern2number(pattern[:-1]) + number
2、N进制
上面方法本质上就是四进制计数,因为一共有4个类型,因此可以用0, 1, 2, 3分别表示碱基A, C, G, T。例如我们枚举0-15序号的串
| AAA 000 00 00 00 |
AAC 001 00 00 01 |
AAG 002 00 00 10 |
AAT 003 00 00 11 |
|---|---|---|---|
| ACA 010 00 01 00 |
ACC 011 00 01 01 |
ACG 012 00 01 01 |
ACT 013 00 01 11 |
| AGA 020 00 10 00 |
AGC 021 00 10 01 |
AGG 022 00 10 10 |
AGT 023 00 10 11 |
| ATA 030 00 11 00 |
ATC 031 00 11 01 |
ATG 032 00 11 10 |
ATT 033 00 11 11 |
2位比特可以表示0-3范围的数,因此如上所示数字越小就表示其字典序越小
symbol2index = dict(zip('ACGT', range(4)))def pattern2number(pattern: str) -> int:digit = 0for ch in pattern:digit = digit * 4 + symbol2index[ch]return digitprint(pattern2number("AGTG"))
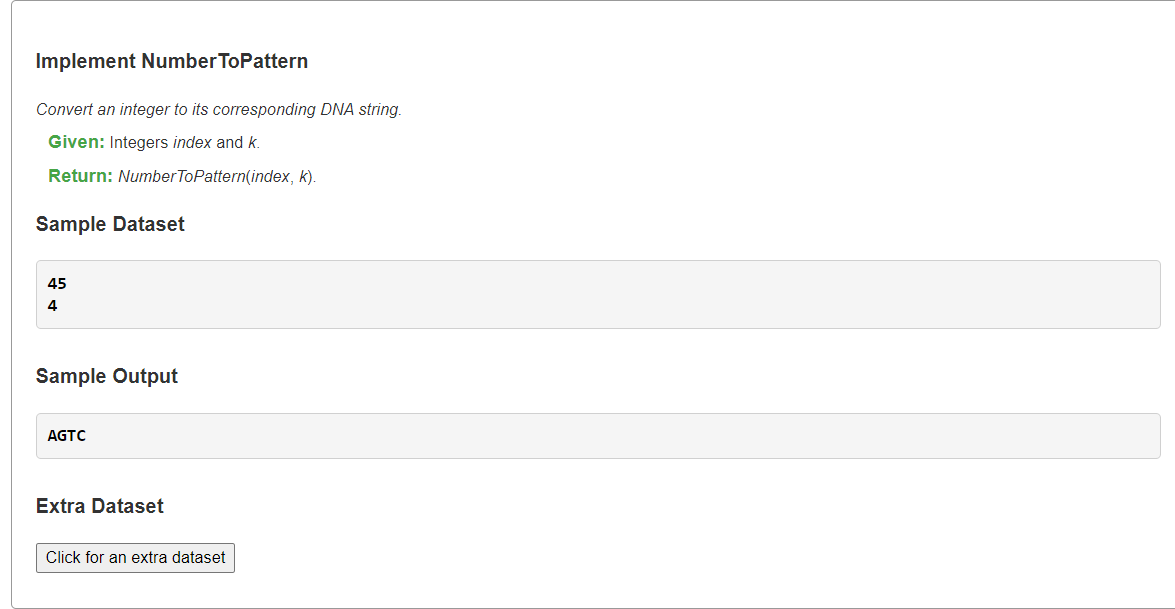
Implement NumberToPattern四进制数转对应字符串
1、从四进制数恢复字符串
上面字符串转四进制数的逆过程,我们可以对四进制数不断%4得最低位的权值,将此权值”翻译”回对应字符,然后不断重复将其/4获得次低位的权值,直到四进制数变为零。
:::info
注意:
题目给定四进制数index和最终字符串长度k的目的是防止index=0时,上述算法就会以为结束,输出位数可能不对。
:::
def number2pattern(index: int, k: int) -> str:# 递归出口if k == 0:return ""# 递归链条prefixIndex, index = divmod(index, 4)return number2pattern(prefixIndex, k - 1) + index2symbol[index]print(number2pattern(45, 4))
def number2pattern(index: int, k: int) -> str:ret = [''] * kwhile k > 0:index, r = divmod(index, 4)ret[k-1] = index2symbol[r] # 逆序填充k -= 1return "".join(ret)print(number2pattern(45, 4))
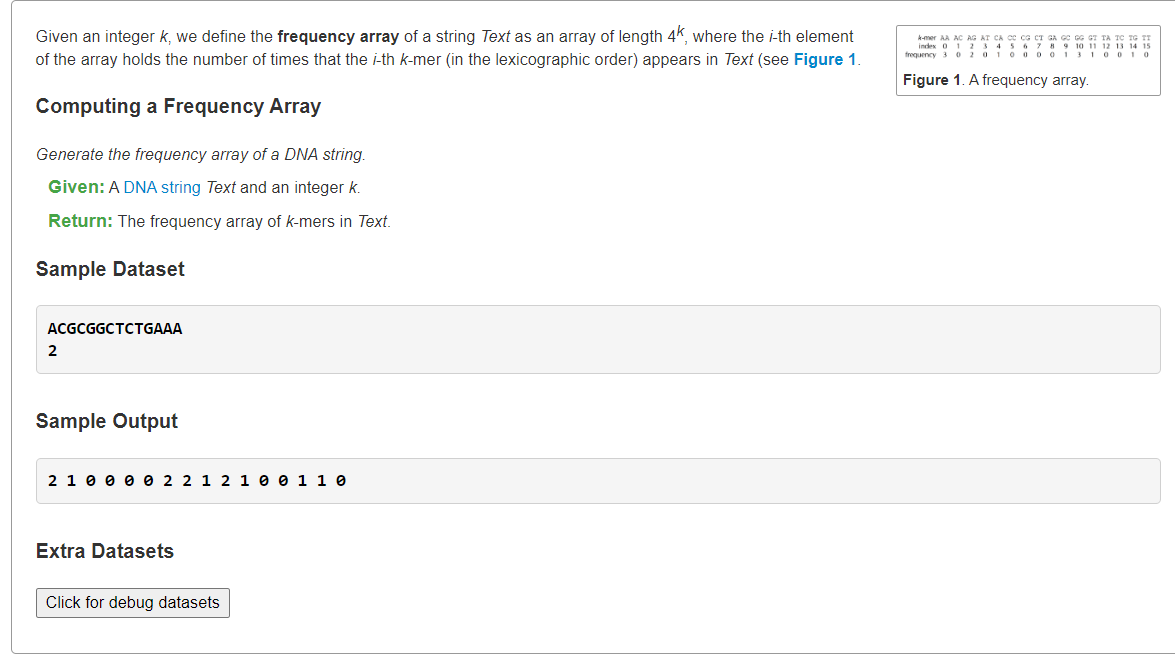
Find the Most Frequent Words with Mismatches in a String4^k个k-mer频数表
1、开足4k哈希桶(枚举所有k-mer串可能情况)
from typing import Listsymbol2int = dict(zip('ACGT', range(4)))def pattern2number(pattern: str) -> int:if not pattern:return 0number = symbol2int[pattern[-1]]return 4 * pattern2number(pattern[:-1]) + numberdef computingFrequencyArray(text: str, k: int) -> List[int]:table = [0] * (4 ** k)for i in range(len(text) - k + 1):table[pattern2number(text[i:i+k])] += 1 # 转为4进制,hasherreturn tables = """ACGCGGCTCTGAAA2"""lines = s.splitlines()text = lines[0]k = int(lines[1])print(*computingFrequencyArray(text, k))
:::info
只是本题要求所有k-mer在Text串中的频数表,这样耗费空间。
:::
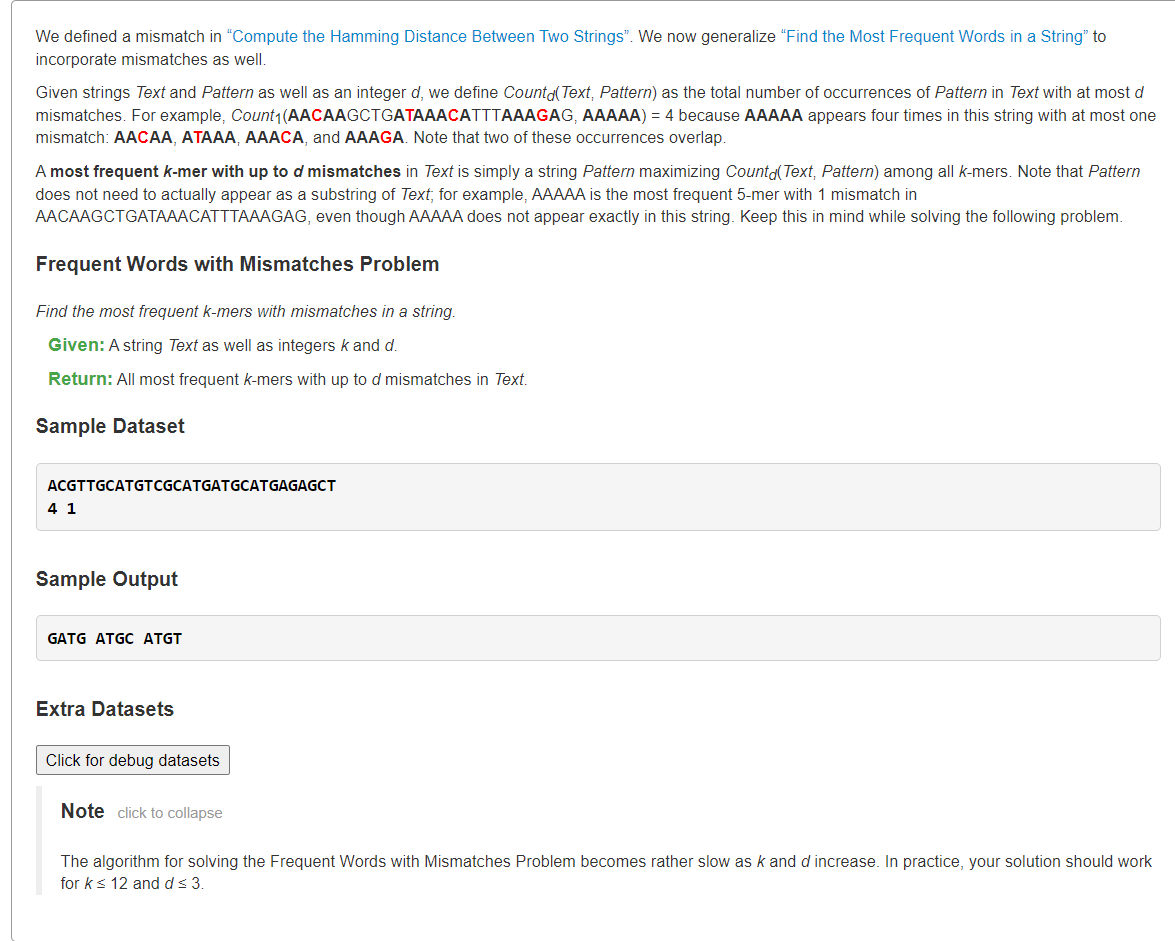
Find the Most Frequent Words with Mismatches in a String4^k个k-mer汉明距离≤d
1、枚举4k个k-mer并用暴力双指针匹配
from typing import Listdef hammingDistance(s: str, t: str) -> int:dist = 0for ch1, ch2 in zip(s, t):if ch1 != ch2:dist += 1return distdef approximatePatternMatch(text: str, pattern: str, d: int) -> List[int]:m, n = len(text), len(pattern)ans = []for i in range(m - n + 1):if hammingDistance(text[i:i+n], pattern) <= d:ans.append(i)return ansdef frequentWordswithMismatches(text: str, k: int, d: int) -> List[str]:def backtrace(n: int, path: List[str], ans: List[str]) -> None:if n == 0:ans.append("".join(path))returnfor base in 'AGCT':path.append(base)backtrace(n - 1, path, ans)path.pop()kmers = []backtrace(k, [], kmers)# print(kmers)ans, maxv = [], 0for pattern in kmers:cnt = len(approximatePatternMatch(text, pattern, d))if cnt > maxv:ans = [pattern]maxv = cntelif cnt == maxv:ans.append(pattern)return anss = """ACGTTGCATGTCGCATGATGCATGAGAGCT4 1"""lines = s.splitlines()text = lines[0]k, d= map(int, lines[1].split())print(*frequentWordswithMismatches(text, k, d))
并行计算:
Find Frequent Words with Mismatches and Reverse Complements