- R
- shiny
survivalsurvminer
- shiny
内置数据
- 甲基化数据:M450K from TCGA GDAC Firehose
- 甲基化水平:β-values (ranging from 0 to 1)
- 甲基化水平:β-values (ranging from 0 to 1)
- 临床数据:survival status, age, sex, height, weight, race, stage and grade of the cancer, etc (与甲基化数据对应)
- cancer type: 25
- 甲基化组:7358
方法
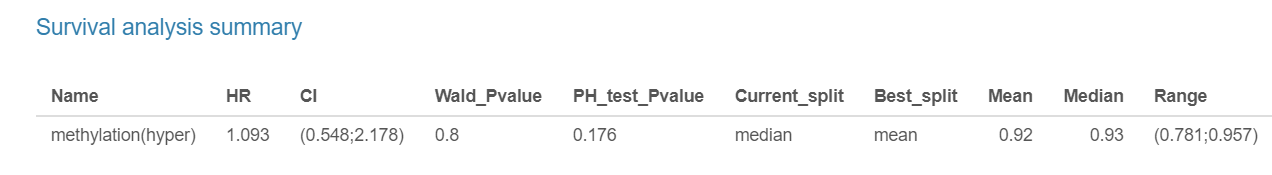
univariable and multivariable Cox regression analysis生存分析(主要关注)
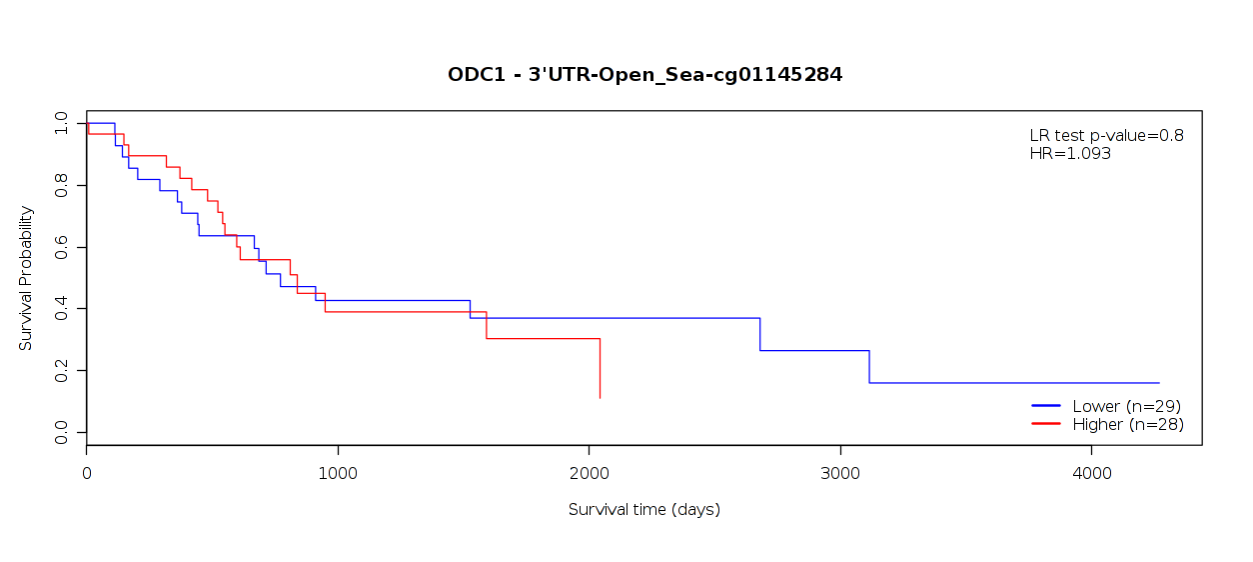
选择基因名后,选择CpG岛、基因组区域、CpG位点、分组方法,可得到KM图。
我的理解是:针对单基因,确定到该基因的CpG岛、基因组区域、CpG位点,用甲基化矩阵做分析。在CpG位点中的胞嘧啶可以被甲基化为5-甲基胞嘧啶。在哺乳动物中,70%到80%的CpG位点的胞嘧啶是甲基化的。https://zh.wikipedia.org/wiki/CpG%E4%BD%8D%E7%82%B9
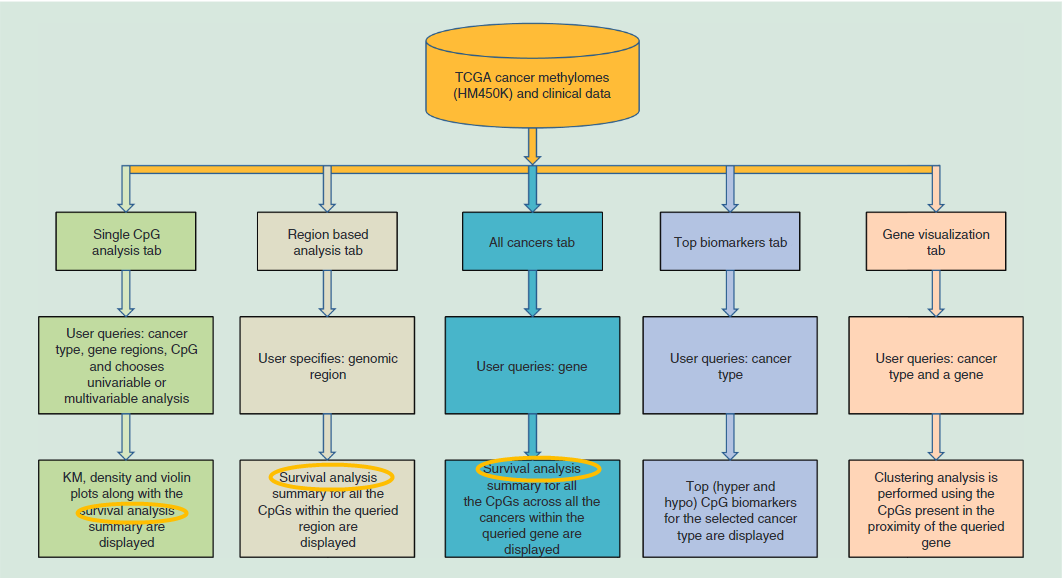
MethSurv enables cancer survival prediction for single CpG sites (‘Single CpG’ analysis tab) using any of the available genes (official gene symbol) in the TCGA HM450K cancer methylome datasets. we provide the users with a detailed overview of individual CpG sites with the options to select genomic regions (relative to CGI and gene sub-region), methods to establish cut-off points for dichotomizing higher and lower methylation patient groups (mean, median, lower quantile, upper quantile and maxstat) and adjustment type.