GTF文件
下载
用于将表达矩阵分为「CDs、NCDs」。
Ensembl http://asia.ensembl.org/info/data/ftp/index.html
→ ftp://ftp.ensembl.org/pub/release-99/gtf/homo_sapiens
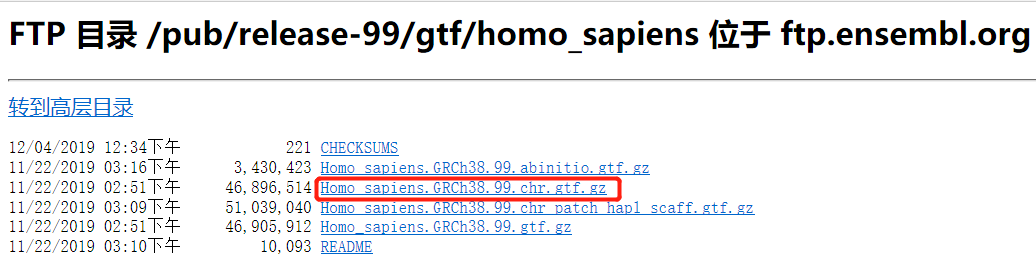
比较神奇的是,chrome居然打不开这个网址(无论怎么清理cookie/换ip)

读入R
直接以” tabular format”读入。
在数据来源和分类里迷惑了半天,后来发现似乎并不需要考虑那么多…
mygtf <- data.table::fread("Homo_sapiens.GRCh38.99.chr.gtf/Homo_sapiens.GRCh38.99.chr.gtf", sep = "\t")dim(mygtf)# [1] 2904340 9class(mygtf)# [1] "data.table" "data.frame"dim(mygtf)# [1] 2904340 9table(mygtf$V2)## ensembl ensembl_havana ensembl_havana_tagene# 235548 690227 95# havana havana_tagene insdc# 1862976 109820 37# mirbase# 5637table(mygtf$V3)## CDS exon five_prime_utr gene# 763375 1376244 153149 60617# Selenocysteine start_codon stop_codon three_prime_utr# 119 87814 80153 155118# transcript# 227751
miRBase is the central online repository for microRNA (miRNA) nomenclature, sequence data, annotation and target prediction. Griffiths-Jones S, Saini H K, van Dongen S, et al. miRBase: tools for microRNA genomics[J]. Nucleic acids research, 2007, 36(suppl_1): D154-D158
mygtf$ensid <- substr(mygtf$V9,10,24)table(duplicated(mygtf$ensid))mygtf <- mygtf[!duplicated(mygtf$ensid),]dim(mygtf)# [1] 60617 10table(mygtf$V3)## gene# 60617row.names(mygtf) <- mygtf$ensidgenename <- strsplit(mygtf$V9,split = ";")start <- Sys.time()for(i in 1:nrow(mygtf)) {mygtf$genename[i] <- strsplit(genename[[i]][3],split = " ")[[1]][3]}end <- Sys.time()end - start# Time difference of 12.65077 minsmygtf$genename <- gsub('\"',"", mygtf$genename, fixed = TRUE)start <- Sys.time()for(i in 1:nrow(mygtf)) {mygtf$genetype[i] <- strsplit(genename[[i]][5],split = " ")[[1]][3]}end <- Sys.time()end - start# Time difference of 8.673628 minsmygtf$genetype <- gsub('\"',"", mygtf$genetype, fixed = TRUE)
将genetype分为两类,genetype含义参考:https://www.gencodegenes.org/pages/biotypes.html
mygtf$group <- ifelse(mygtf$genetype=="IG_C_gene"|mygtf$genetype=="IG_D_gene"|mygtf$genetype=="IG_J_gene"|mygtf$genetype=="IG_V_gene"|mygtf$genetype=="protein_coding"|mygtf$genetype=="TEC"|mygtf$genetype=="TR_C_gene"|mygtf$genetype=="TR_D_gene"|mygtf$genetype=="TR_J_gene"|mygtf$genetype=="TR_V_gene","CDS","other")table(mygtf$group)## CDS other# 21407 39210cdsid <- mygtf[mygtf$group=="CDS",]$ensidotherid <- mygtf[mygtf$group=="other",]$ensidsave(cdsid,file = "cdsid.Rdata")save(otherid,file = "otherid.Rdata")save(mygtf,file = "humangtf.Rdata")
将TCGA表达矩阵分为两个矩阵存为Rdata
选用 GDC-TCGA 33种癌症 https://xenabrowser.net/datapages/
path <- "../13genes-signature/GDC-TCGA-data"filenames <- list.files(path,pattern = ".tsv")filePath <- sapply(filenames, function(x){paste(path,x,x,sep='/')})filePath <- filePath[c(TRUE, FALSE)]start <- Sys.time()expdata <- lapply(filePath, function(x){expr <- read.table(x,header = T,sep='\t')expr <- expr[1:(nrow(expr)-5),]expr[,1] <- as.character(expr[,1])for (j in 1:nrow(expr)) {expr[j,1] <- strsplit(expr[j,1],"\\.")[[1]][1]}row.names(expr) <- expr[,1]expr <- expr[,-1]expr <- log2(edgeR::cpm(expr)+1)expr <- na.omit(expr)})end <- Sys.time()end - start# Time difference of 37.246043 minsstart <- Sys.time()for(i in 1:length(expdata)){expr <- expdata[[i]]cds_expr <- expr[intersect(row.names(expr),cdsid),]other_expr <- expr[intersect(row.names(expr),otherid),]save(cds_expr,file = paste0(sub("-","",stringr::str_extract(string = names(filePath)[i],pattern = '-\\w*')),"_cds_expr.Rdata"))save(other_expr,file = paste0(sub("-","",stringr::str_extract(string = names(filePath)[i],pattern = '-\\w*')),"_other_expr.Rdata"))}end <- Sys.time()end - start# Time difference of 3.861098 mins
得到了66个拆分后的表达矩阵.Rdata
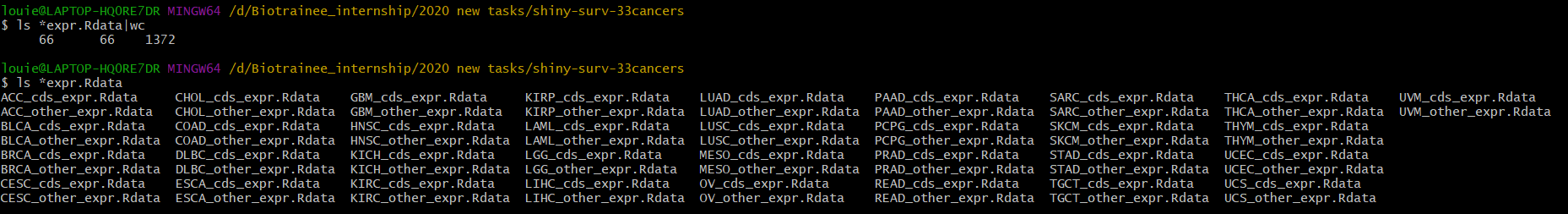
生存数据存为Rdata
path <- "../13genes-signature/GDC-TCGA-data"filenames <- list.files(path,pattern = ".tsv")filePath <- sapply(filenames, function(x){paste(path,x,x,sep='/')})filePath <- filePath[c(FALSE, TRUE)]start <- Sys.time()survdata <- lapply(filePath, function(x){phe <- read.table(x,header = T,sep='\t',fill = T,quote = "")phe$sample = gsub('-','.',phe$sample)row.names(phe) <- phe[,1]return(phe)# id <- intersect(colnames(expr),rownames(phe))# expr <- expr[,id]# phe <- phe[id,]})end <- Sys.time()end - start# Time difference of 0.5615041 secsstart <- Sys.time()for(i in 1:length(survdata)){surv <- survdata[[i]]save(surv,file = paste0(sub("-","",stringr::str_extract(string = names(filePath)[i],pattern = '-\\w*')),"_surv.Rdata"))}end <- Sys.time()end - start# Time difference of 0.2563121 secs

