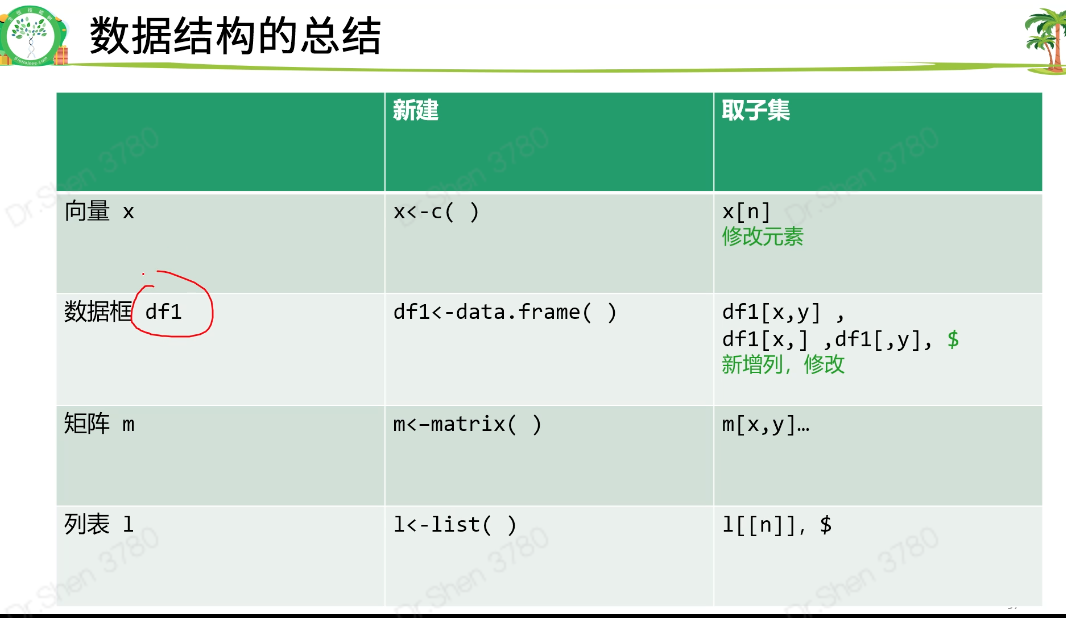
| vector,向量,一维 |
|
| matrix,表格,矩阵,二维, |
只能有1种数据类型 |
| data.frame数据框,二维, |
每列只能有1种数据类型 |
| list列表,包罗万象 |
判断依据:1、生成的函数;2、class或is函数
数据框来源
- 代码新建
- 已有数据转换或处理结果
- 读取表格文件
- R语言内置数据,eg.LETTERS
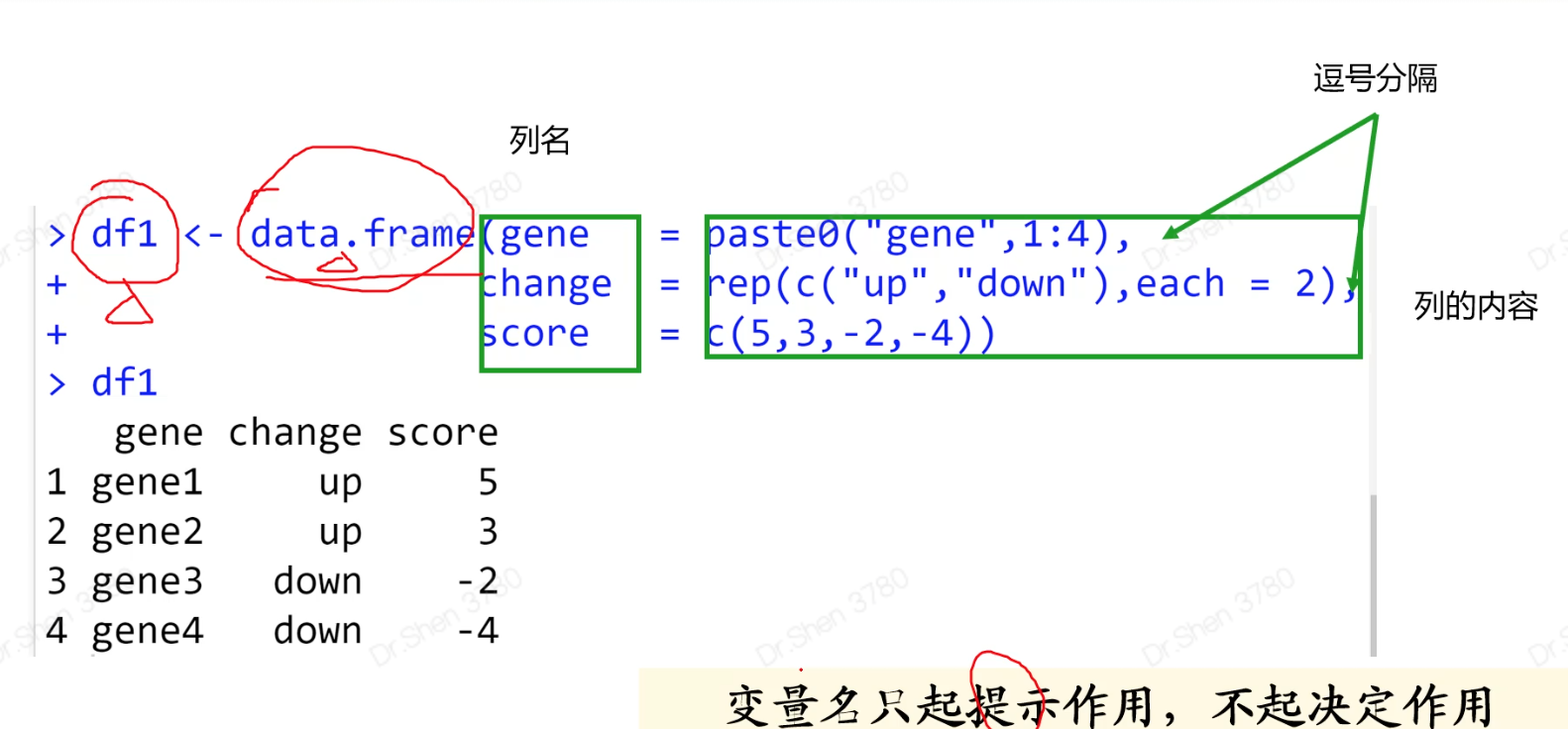
新建数据框
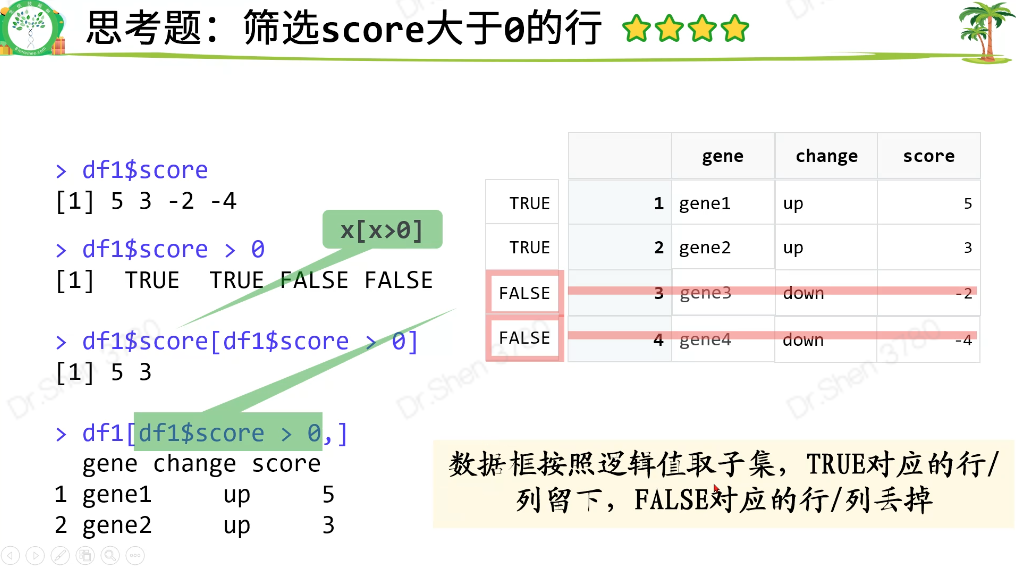
筛选score>0 的行
删除变量

> #重点:数据框> #1.数据框来源> # (1)用代码新建> # (2)由已有数据转换或处理得到> # (3)读取表格文件> # (4)R语言内置数据> > #2.新建和读取数据框> df1 <- data.frame(gene = paste0("gene",1:4),+ change = rep(c("up","down"),each = 2),+ score = c(5,3,-2,-4))> df1 gene change score1 gene1 up 52 gene2 up 33 gene3 down -24 gene4 down -4> > df2 <- read.csv("gene.csv")> df2 gene change score1 gene1 up 52 gene2 up 33 gene3 down -24 gene4 down -4> > #3.数据框属性> #维度,指行数和列数> dim(df1)[1] 4 3> nrow(df1)#行数[1] 4> ncol(df1)#列数[1] 3> #行名和列名> rownames(df1)[1] "1" "2" "3" "4"> colnames(df1)[1] "gene" "change" "score" > > #4.数据框取子集> df1$gene # $一次只能提取1列,不能多列[1] "gene1" "gene2" "gene3" "gene4"> df1$score#tab键可协助补齐[1] 5 3 -2 -4> mean(df1$score)[1] 0.5> > ## 按坐标> df1[2,2][1] "up"> df1[2,] #取第二行 gene change score2 gene2 up 3> df1[,2] #取第二列[1] "up" "up" "down" "down"> df1[c(1,3),1:2] #第一、三行和第一、二列 gene change1 gene1 up3 gene3 down> > ## 按名字> df1[,"gene"] #可实现1次提取多列[1] "gene1" "gene2" "gene3" "gene4"> df1[,c('gene','change')] gene change1 gene1 up2 gene2 up3 gene3 down4 gene4 down> > ## 按条件(逻辑值)> df1[df1$score>0,]#筛选score>0的行 gene change score1 gene1 up 52 gene2 up 3> #拆分解答> df1$score[1] 5 3 -2 -4> df1$score>0[1] TRUE TRUE FALSE FALSE> df1$score[df1$score>0][1] 5 3> df1[df1$score>0,] gene change score1 gene1 up 52 gene2 up 3> > df1[df1$score>0,1]#筛选score>0的基因[1] "gene1" "gene2"> df1$gene[df1$score>0]#筛选score>0的基因[1] "gene1" "gene2"> > #5.数据框修改> > #改一个格> df1[3,3] <- 5> df1 gene CHANGE score p.valuer1 gene1 up 12 0.01r2 gene2 up 23 0.02r3 gene3 down 5 0.07r4 gene4 down 2 0.05> #改一整列> df1$score <- c(12,23,50,2) > df1 gene CHANGE score p.valuer1 gene1 up 12 0.01r2 gene2 up 23 0.02r3 gene3 down 50 0.07r4 gene4 down 2 0.05> #新增一列> df1$p.value <- c(0.01,0.02,0.07,0.05) #对新的名称而言是新增,对原有的而言是修改> df1 gene CHANGE score p.valuer1 gene1 up 12 0.01r2 gene2 up 23 0.02r3 gene3 down 50 0.07r4 gene4 down 2 0.05> > #改行名和列名> rownames(df1) <- c("r1","r2","r3","r4")> #只修改某一行/列的名> colnames(df1)[2] <- "CHANGE"> > #6.两个数据框的连接,merge左连接、右连接、取交集> test1 <- data.frame(name = c('jimmy','nicker','Damon','Sophie'), + blood_type = c("A","B","O","AB"))> test1 name blood_type1 jimmy A2 nicker B3 Damon O4 Sophie AB> test2 <- data.frame(name = c('Damon','jimmy','nicker','tony'),+ group = c("group1","group1","group2","group2"),+ vision = c(4.2,4.3,4.9,4.5))> test2 name group vision1 Damon group1 4.22 jimmy group1 4.33 nicker group2 4.94 tony group2 4.5> > test3 <- data.frame(NAME = c('Damon','jimmy','nicker','tony'),+ weight = c(140,145,110,138))> test3 NAME weight1 Damon 1402 jimmy 1453 nicker 1104 tony 138> merge(test1,test2,by="name") name blood_type group vision1 Damon O group1 4.22 jimmy A group1 4.33 nicker B group2 4.9> merge(test1,test3,by.x = "name",by.y = "NAME")#用于名称的大小写字母不一致时 name blood_type weight1 Damon O 1402 jimmy A 1453 nicker B 110> > ##### 矩阵和列表> m <- matrix(1:9, nrow = 3)> colnames(m) <- c("a","b","c") #加列名> m a b c[1,] 1 4 7[2,] 2 5 8[3,] 3 6 9> #取子集,不支持$> m[2,]a b c 2 5 8 > m[,1][1] 1 2 3> m[2,3]c 8 > m[2:3,1:2] a b[1,] 2 5[2,] 3 6> m a b c[1,] 1 4 7[2,] 2 5 8[3,] 3 6 9> t(m) #转置 [,1] [,2] [,3]a 1 2 3b 4 5 6c 7 8 9> as.data.frame(m) # 需要再赋值才会变 a b c1 1 4 72 2 5 83 3 6 9> > #列表,列表的下一级是元素> l <- list(m1 = matrix(1:9, nrow = 3),+ m2 = matrix(2:9, nrow = 2))> l$m1 [,1] [,2] [,3][1,] 1 4 7[2,] 2 5 8[3,] 3 6 9$m2 [,1] [,2] [,3] [,4][1,] 2 4 6 8[2,] 3 5 7 9> > l[[2]]#取子集,取l列表中的第2个元素 [,1] [,2] [,3] [,4][1,] 2 4 6 8[2,] 3 5 7 9> l$m1 # 取l列表中的m1元素 [,1] [,2] [,3][1,] 1 4 7[2,] 2 5 8[3,] 3 6 9> > # 补充:元素的名字> scores = c(100,59,73,95,45)> names(scores) = c("jimmy","nicker","Damon","Sophie","tony")> scores jimmy nicker Damon Sophie tony 100 59 73 95 45 > scores["jimmy"]jimmy 100 > scores[c("jimmy","nicker")] jimmy nicker 100 59 > > names(scores)[scores>60] #选出>60的子集[1] "jimmy" "Damon" "Sophie"> > # 删除变量 > rm(l) #删除一个> rm(df1,df2) #删除多个Warning messages:1: In rm(df1, df2) : 找不到对象'df1'2: In rm(df1, df2) : 找不到对象'df2'> rm(list = ls()) #删除全部> #清空控制台: ctrl+l> > #调整元素顺序> x <- c("A","B","C","D","E");x[1] "A" "B" "C" "D" "E"> x[c(2,4,1,3,5)][1] "B" "D" "A" "C" "E"> > scores=c(100,59,73,95,45);scores[1] 100 59 73 95 45> scores[c(5,2,3,4,1)][1] 45 59 73 95 100> sort(scores) #另一种方式,从小到大排序[1] 45 59 73 95 100> order(scores) #通过order取子集生成的结果等同于sort[1] 5 2 3 4 1> > #向量匹配排序,match> x <- c("A","B","C","D","E")> y <- c("B","D","A","C","E")> match(y,x)#以y为模版、目标、结果,以x为原料,去进行调整顺序所得到的下标[1] 2 4 1 3 5> x[match(y,x)][1] "B" "D" "A" "C" "E"> 练习> # 练习3-2> # 1.统计内置数据iris最后一列有哪几个取值,每个取值重复了多少次> iris[,ncol(iris)] [1] setosa setosa setosa setosa [5] setosa setosa setosa setosa [9] setosa setosa setosa setosa [13] setosa setosa setosa setosa [17] setosa setosa setosa setosa [21] setosa setosa setosa setosa [25] setosa setosa setosa setosa [29] setosa setosa setosa setosa [33] setosa setosa setosa setosa [37] setosa setosa setosa setosa [41] setosa setosa setosa setosa [45] setosa setosa setosa setosa [49] setosa setosa versicolor versicolor [53] versicolor versicolor versicolor versicolor [57] versicolor versicolor versicolor versicolor [61] versicolor versicolor versicolor versicolor [65] versicolor versicolor versicolor versicolor [69] versicolor versicolor versicolor versicolor [73] versicolor versicolor versicolor versicolor [77] versicolor versicolor versicolor versicolor [81] versicolor versicolor versicolor versicolor [85] versicolor versicolor versicolor versicolor [89] versicolor versicolor versicolor versicolor [93] versicolor versicolor versicolor versicolor [97] versicolor versicolor versicolor versicolor[101] virginica virginica virginica virginica [105] virginica virginica virginica virginica [109] virginica virginica virginica virginica [113] virginica virginica virginica virginica [117] virginica virginica virginica virginica [121] virginica virginica virginica virginica [125] virginica virginica virginica virginica [129] virginica virginica virginica virginica [133] virginica virginica virginica virginica [137] virginica virginica virginica virginica [141] virginica virginica virginica virginica [145] virginica virginica virginica virginica [149] virginica virginica Levels: setosa versicolor virginica> table(iris[,ncol(iris)]) setosa versicolor virginica 50 50 50 > > # 2.提取内置数据iris的前5行,前4列,并转换为矩阵,赋值给a。> iris[1:5,1:4] Sepal.Length Sepal.Width Petal.Length Petal.Width1 5.1 3.5 1.4 0.22 4.9 3.0 1.4 0.23 4.7 3.2 1.3 0.24 4.6 3.1 1.5 0.25 5.0 3.6 1.4 0.2> a <- as.matrix(iris[1:5,1:4])> a Sepal.Length Sepal.Width Petal.Length Petal.Width1 5.1 3.5 1.4 0.22 4.9 3.0 1.4 0.23 4.7 3.2 1.3 0.24 4.6 3.1 1.5 0.25 5.0 3.6 1.4 0.2> > # 3.将a的行名改为f lower1,flower2...flower5。> row.names(a) <- paste0("flower",1:5)> row.names(a) <- paste0("flower",1:nrow(a))> a Sepal.Length Sepal.Width Petal.Lengthflower1 5.1 3.5 1.4flower2 4.9 3.0 1.4flower3 4.7 3.2 1.3flower4 4.6 3.1 1.5flower5 5.0 3.6 1.4 Petal.Widthflower1 0.2flower2 0.2flower3 0.2flower4 0.2flower5 0.2> > # 4.探索列表取子集l[2]和l[[2]]的区别(提示:数据结构)> l <- list(m1 = matrix(1:9, nrow = 3),+ m2 = matrix(2:9, nrow = 2))> l$m1 [,1] [,2] [,3][1,] 1 4 7[2,] 2 5 8[3,] 3 6 9$m2 [,1] [,2] [,3] [,4][1,] 2 4 6 8[2,] 3 5 7 9> l[2]$m2 [,1] [,2] [,3] [,4][1,] 2 4 6 8[2,] 3 5 7 9> l[[2]] [,1] [,2] [,3] [,4][1,] 2 4 6 8[2,] 3 5 7 9> class(l[2])#列表,且列表中只有1个矩阵[1] "list"> class(l[[2]])#取子集,不带列表[1] "matrix" "array" > match函数的使用> load("matchtest.Rdata")> #a和b是两个内容相同大顺序不同的向量,才用match> #a> x$file_name[1] "708a16a3-7a5e-4e27-b06b-4c3c308b11fe.htseq.counts.gz"[2] "95e726db-5ccc-4836-a2ae-7feaddaf9f1b.htseq.counts.gz"[3] "90a46dce-5762-47ec-925c-deff853069aa.htseq.counts.gz"[4] "587e44e4-87ba-4981-a520-d20612486f53.htseq.counts.gz"[5] "1b843dbb-5ef0-47ca-9783-dbeb94aa6df3.htseq.counts.gz"[6] "09796233-3f40-4deb-b77d-2267c3afff59.htseq.counts.gz"[7] "44f1dc34-a01e-4a7b-a7a1-a90064039fdd.htseq.counts.gz"> #b> colnames(y)[1] "90a46dce-5762-47ec-925c-deff853069aa.htseq.counts.gz"[2] "587e44e4-87ba-4981-a520-d20612486f53.htseq.counts.gz"[3] "95e726db-5ccc-4836-a2ae-7feaddaf9f1b.htseq.counts.gz"[4] "09796233-3f40-4deb-b77d-2267c3afff59.htseq.counts.gz"[5] "708a16a3-7a5e-4e27-b06b-4c3c308b11fe.htseq.counts.gz"[6] "44f1dc34-a01e-4a7b-a7a1-a90064039fdd.htseq.counts.gz"[7] "1b843dbb-5ef0-47ca-9783-dbeb94aa6df3.htseq.counts.gz"> #a %in% b,核查是否内容相同> table(x$file_name) %in% colnames(y)[1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE> table(colnames(y) %in% x$file_name)TRUE 7 > #a[match(b,a)]> m=x$file_name[match(colnames(y),x$file_name)]> m==colnames(y) #检查前后二者是否一致[1] TRUE TRUE TRUE TRUE TRUE TRUE TRUE> identical(m,colnames(y))#检查前后二者是否一致[1] TRUE> #属于x$file_name的下标,也可以给x$ID用,因为对应> #所以match(colnames(y),x$file_name)也可以给x$ID用> n = x$ID[match(colnames(y),x$file_name)]> #11行和16行的两列按照相同下标子集> #对于本来对应的额,取完子集仍对应;因此m和n对应,13行和colnames(y)对应> colnames(y)=n> > #方法2:调整x行的顺序,让它和colnames(y)对应> ??错误: unexpected input在"?"里> > #方法3:调整y行的顺序,让它和x$file_name对应> ???错误: unexpected input在"?"里>





