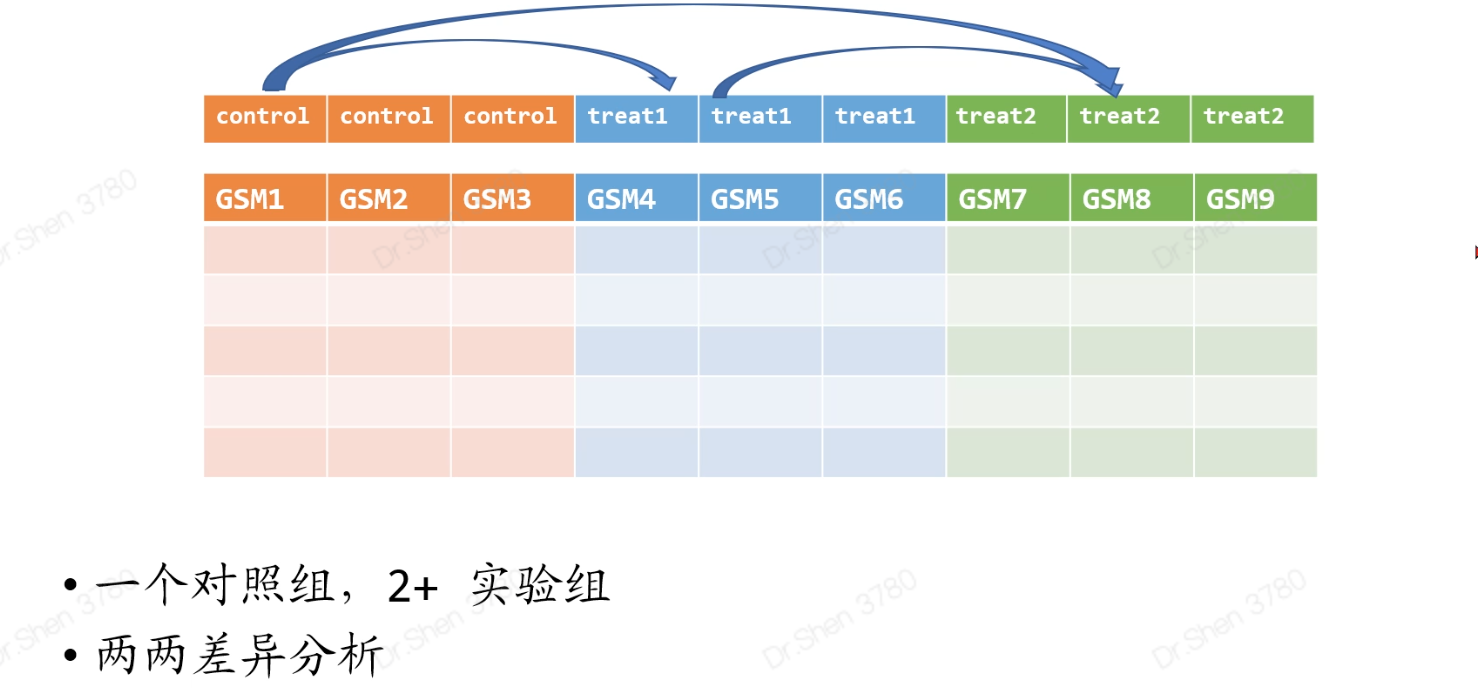
多分组数据
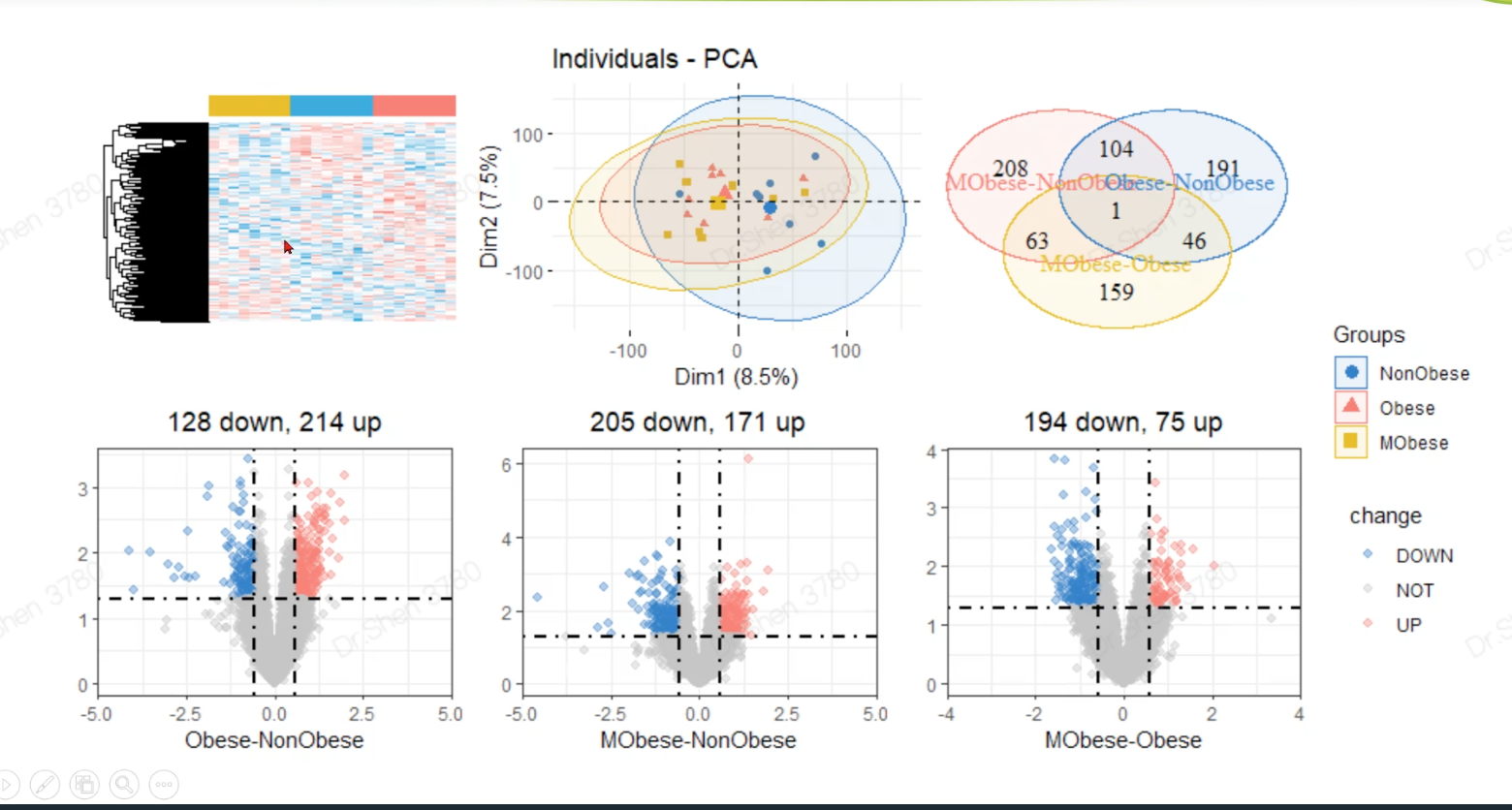
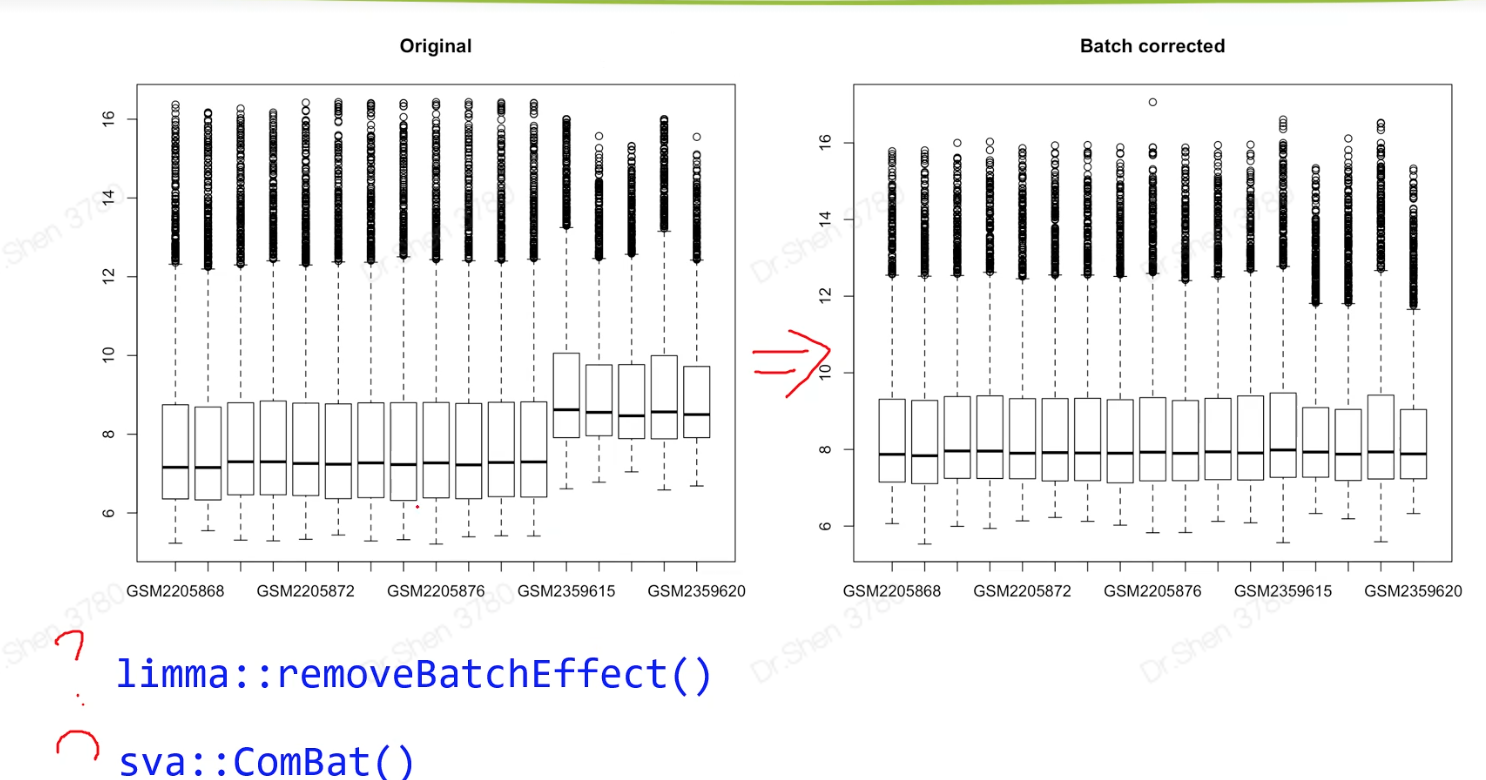
多数据联合分析


标准流程后续分析

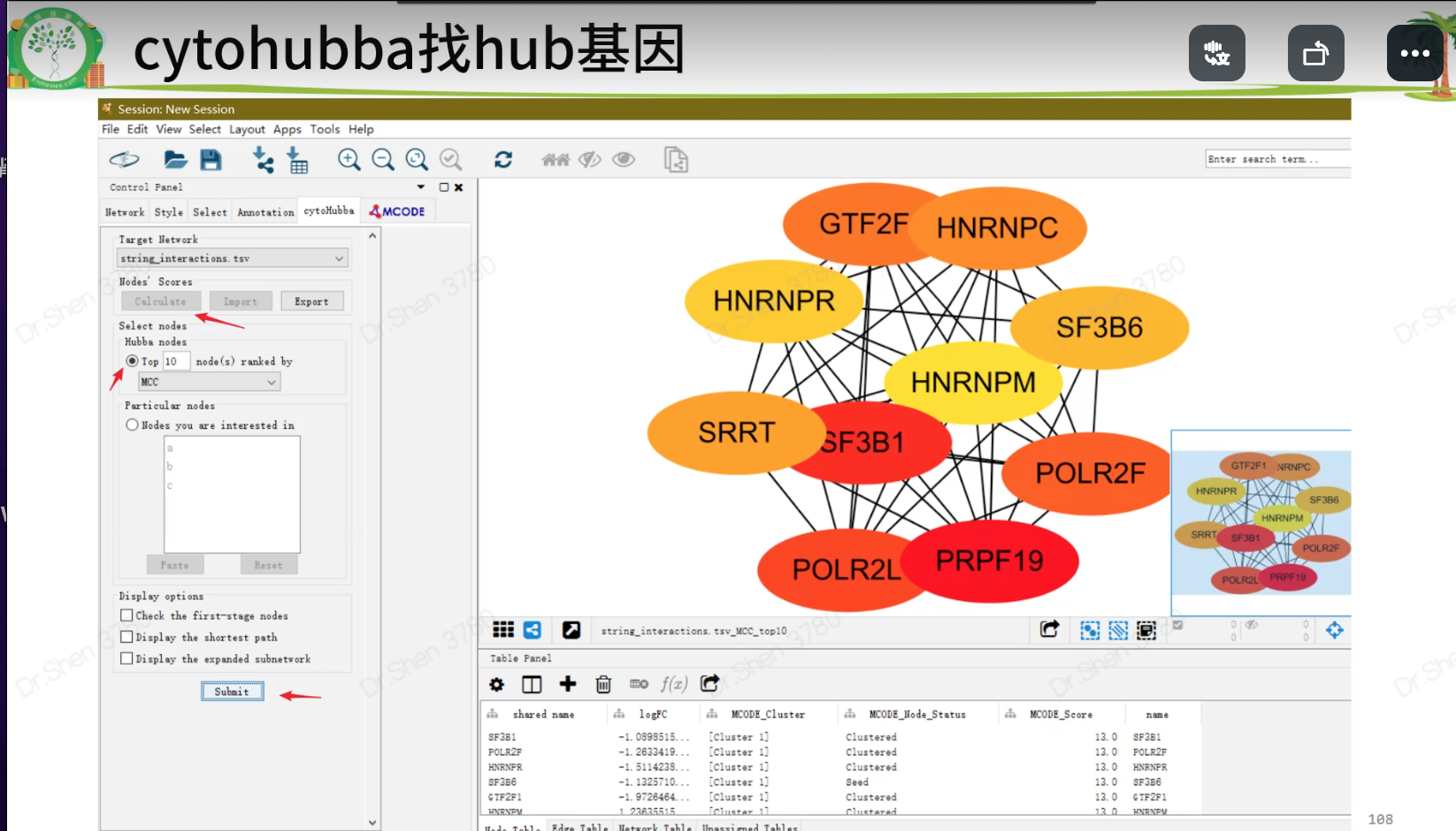
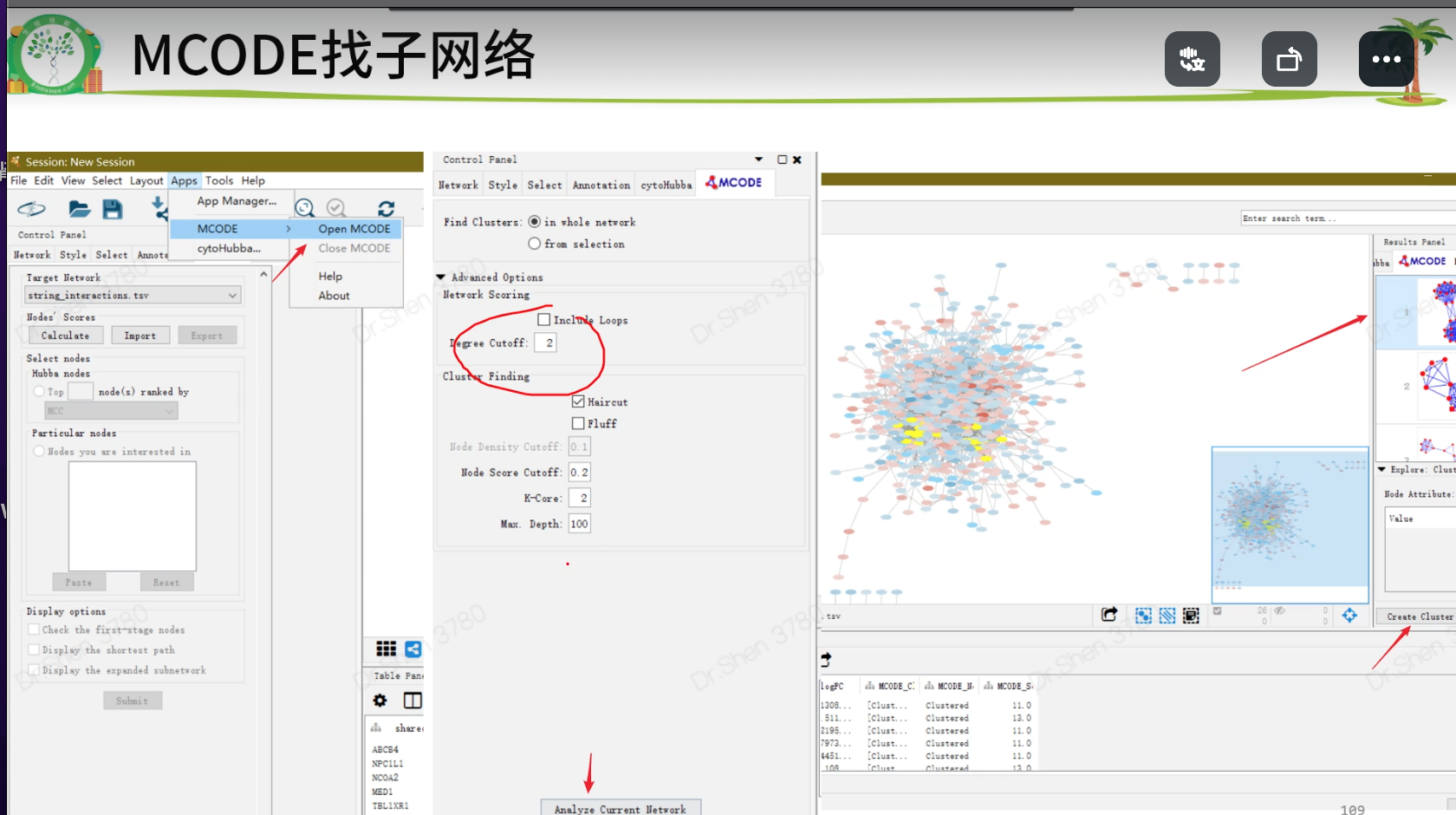
string、cytoscape
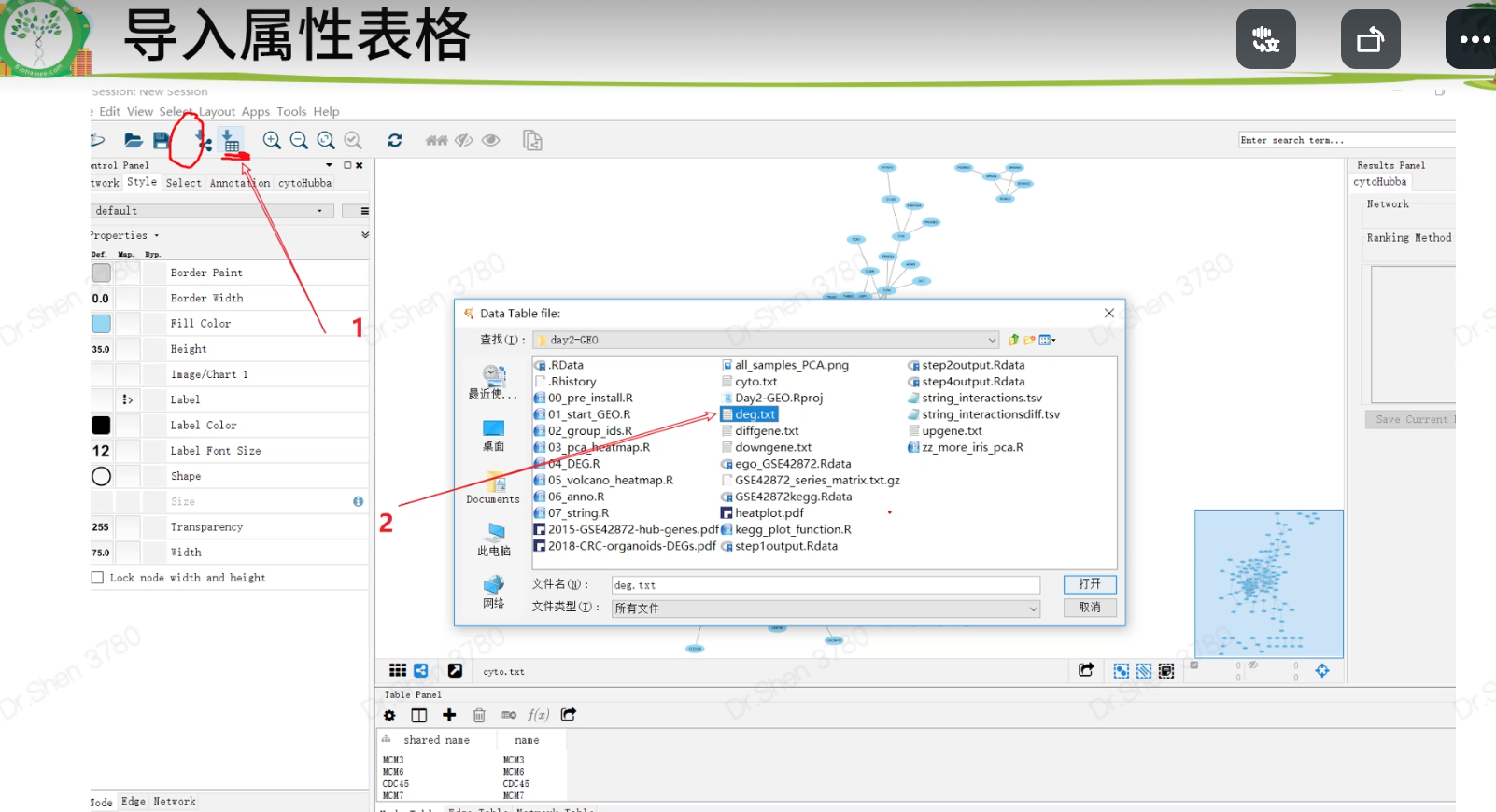
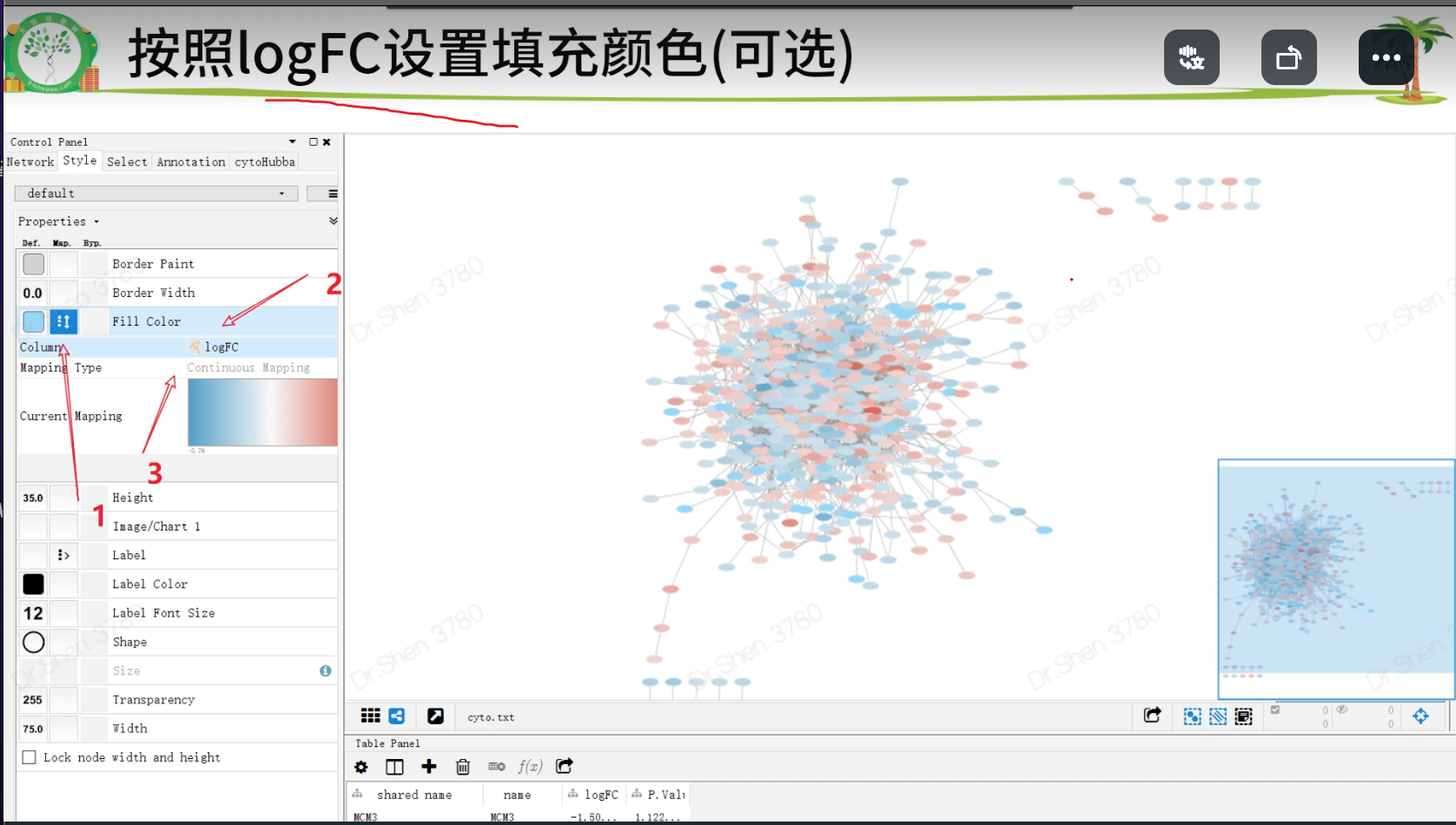
rm(list = ls()) #清空前期load("step4output.Rdata")gene_up= deg[deg$change == 'up','symbol'] gene_down=deg[deg$change == 'down','symbol']gene_diff = c(gene_up,gene_down)# 1.制作string的输入数据write.table(gene_diff, file="diffgene.txt", row.names = F, col.names = F, quote = F)# 从string网页获得string_interactions.tsv# 2.准备cytoscape的输入文件p = deg[deg$change != "stable", c("symbol","logFC")]head(p)write.table(p, file = "deg.txt", sep = "\t", quote = F, row.names = F)# string_interactions.tsv是网络文件# deg.txt是属性表格
文献解读
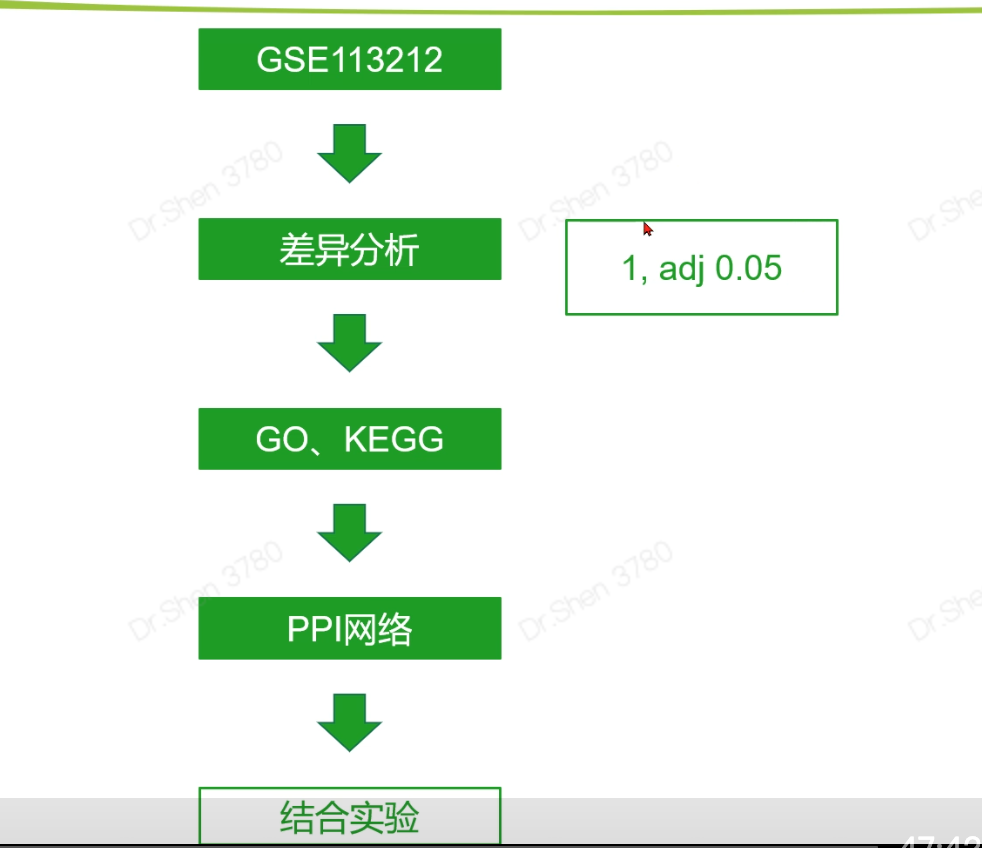
文献1 二分组

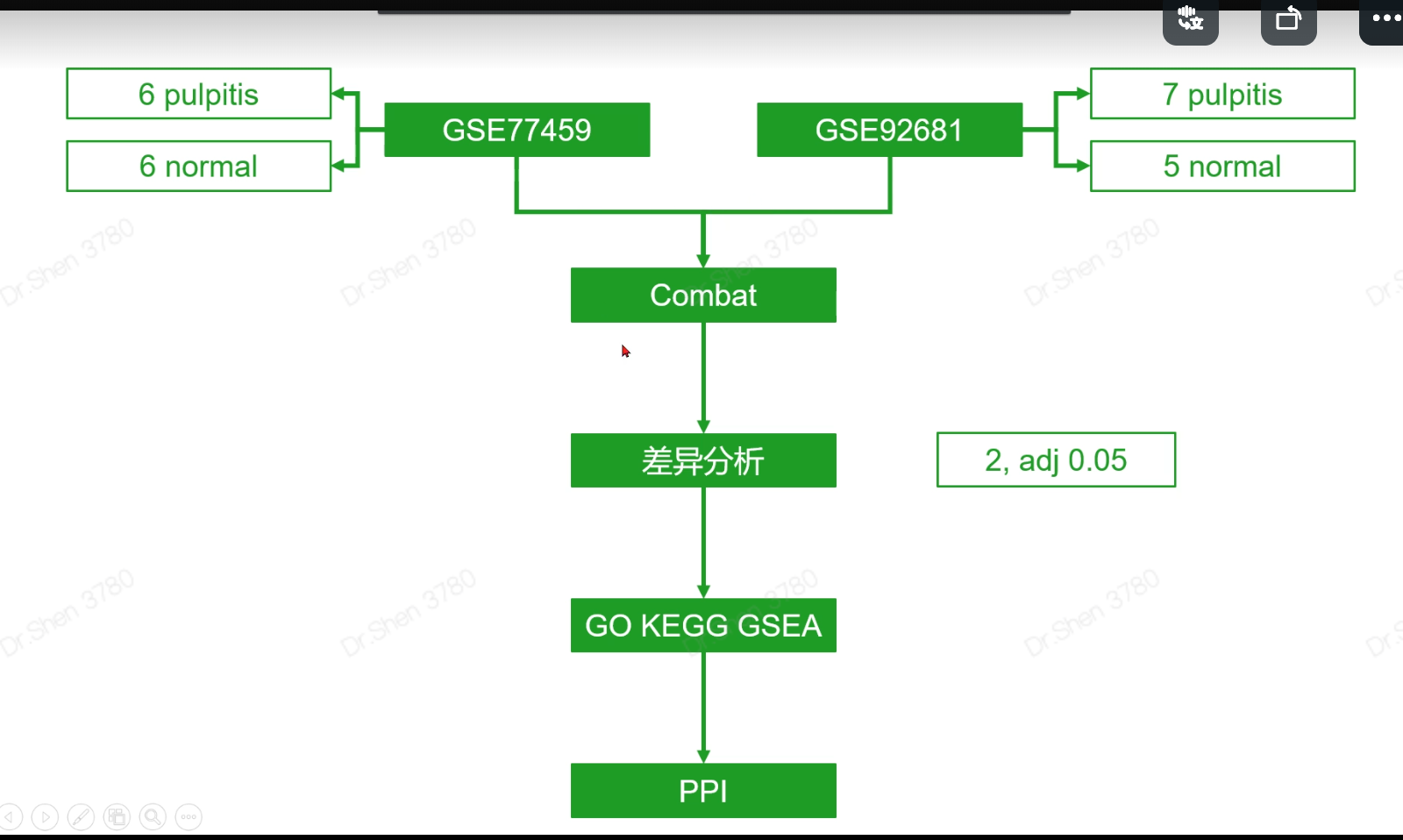
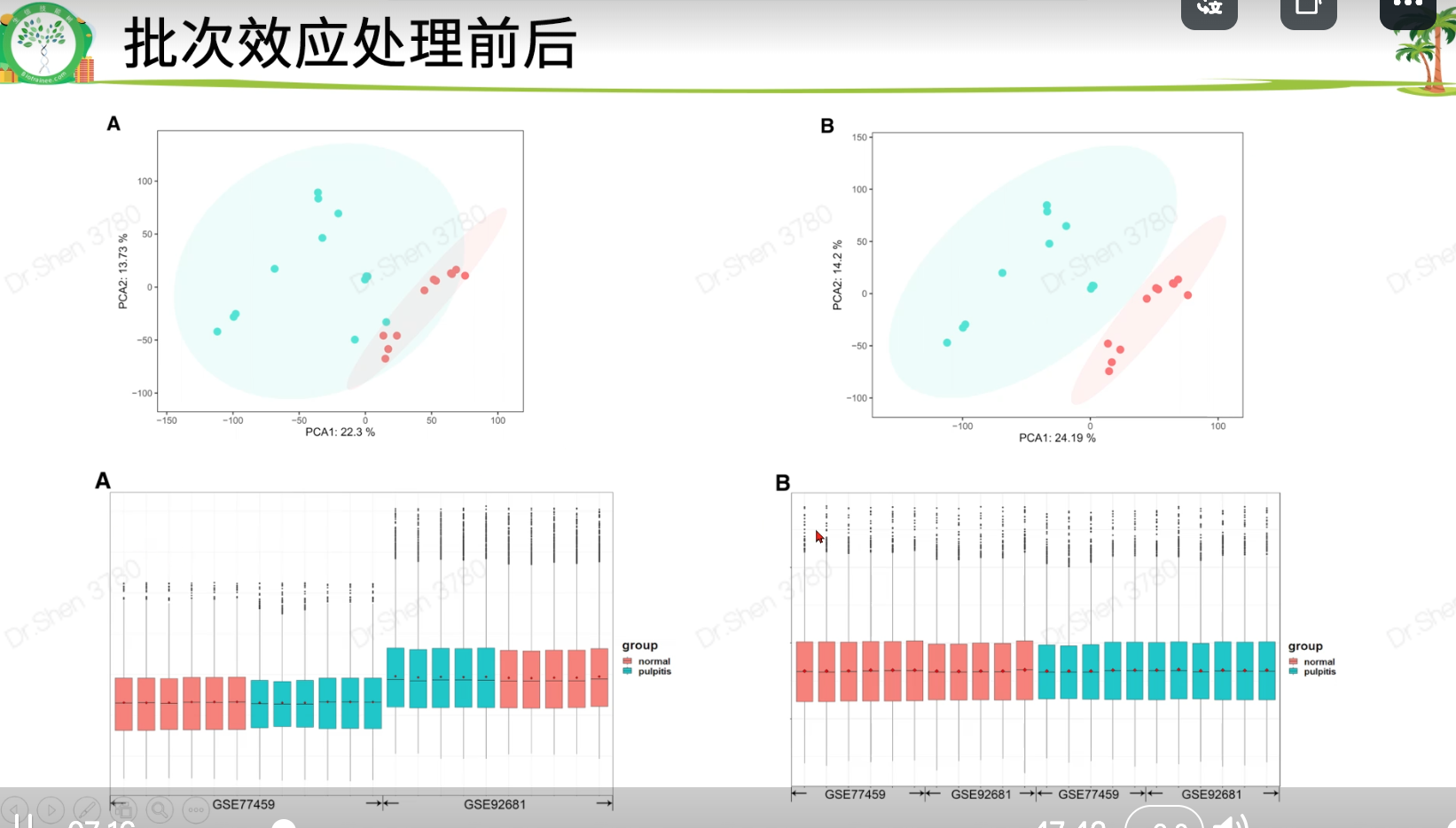
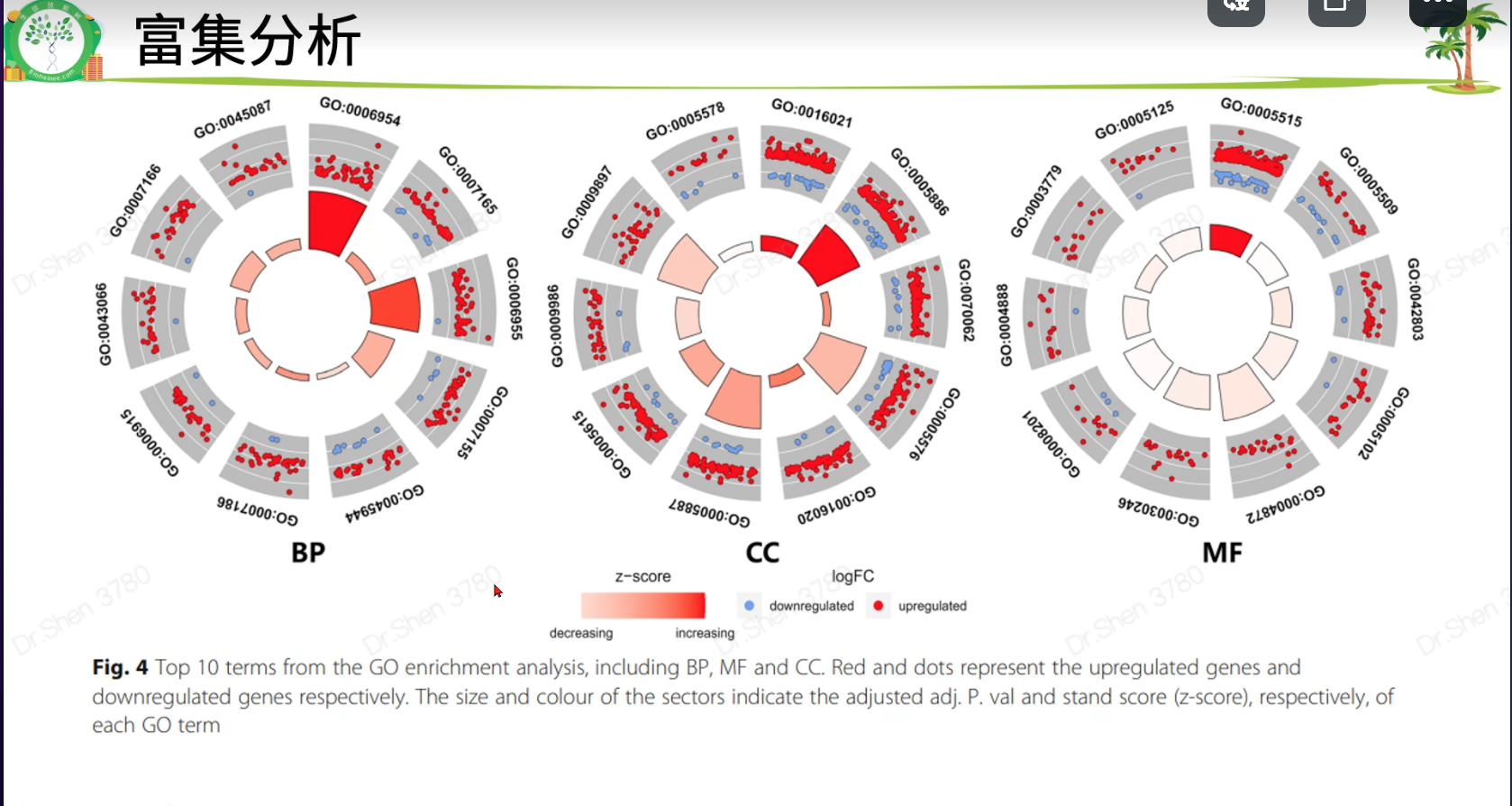
文献2 多数据

文献3 转录因子 多数据

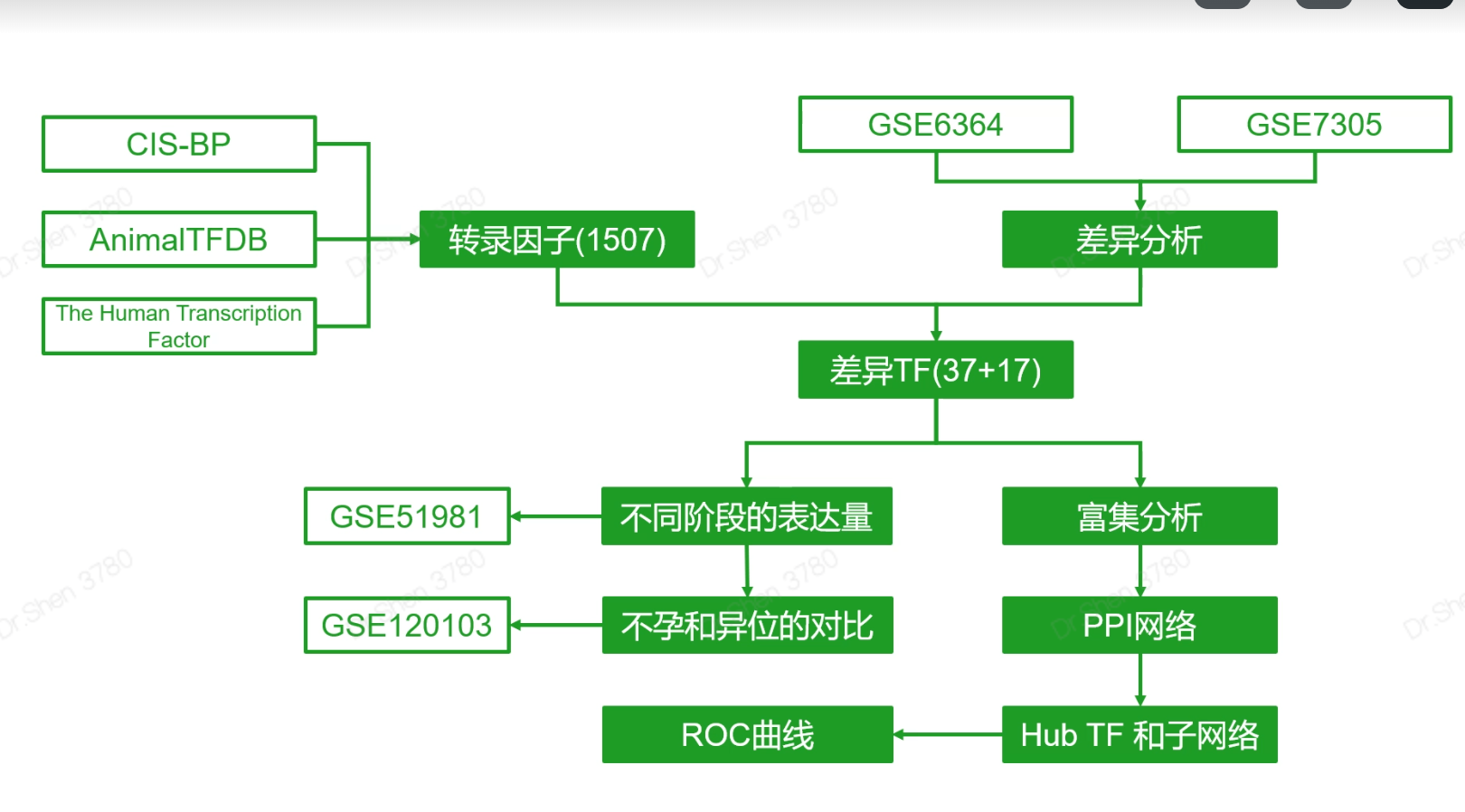
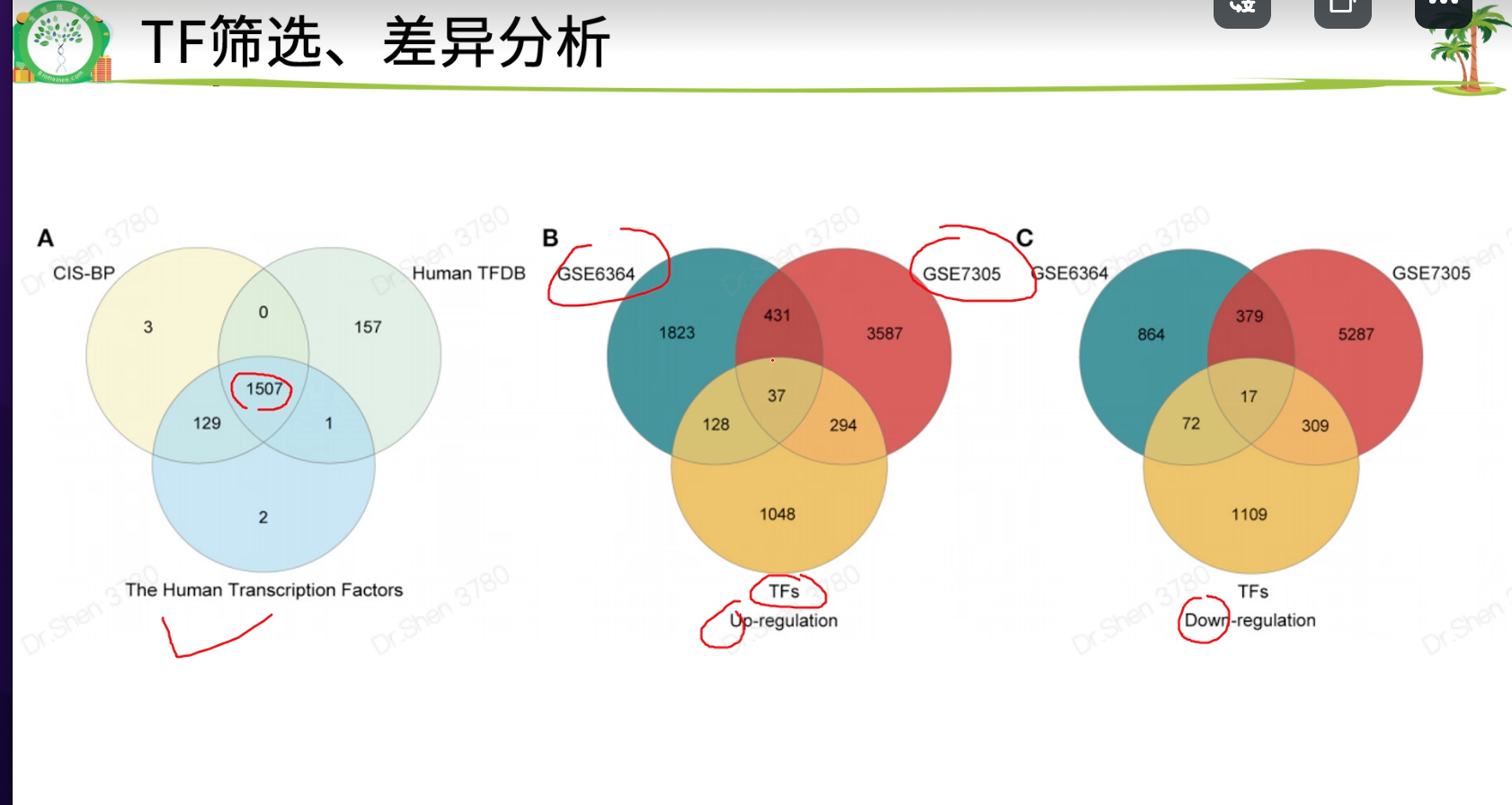
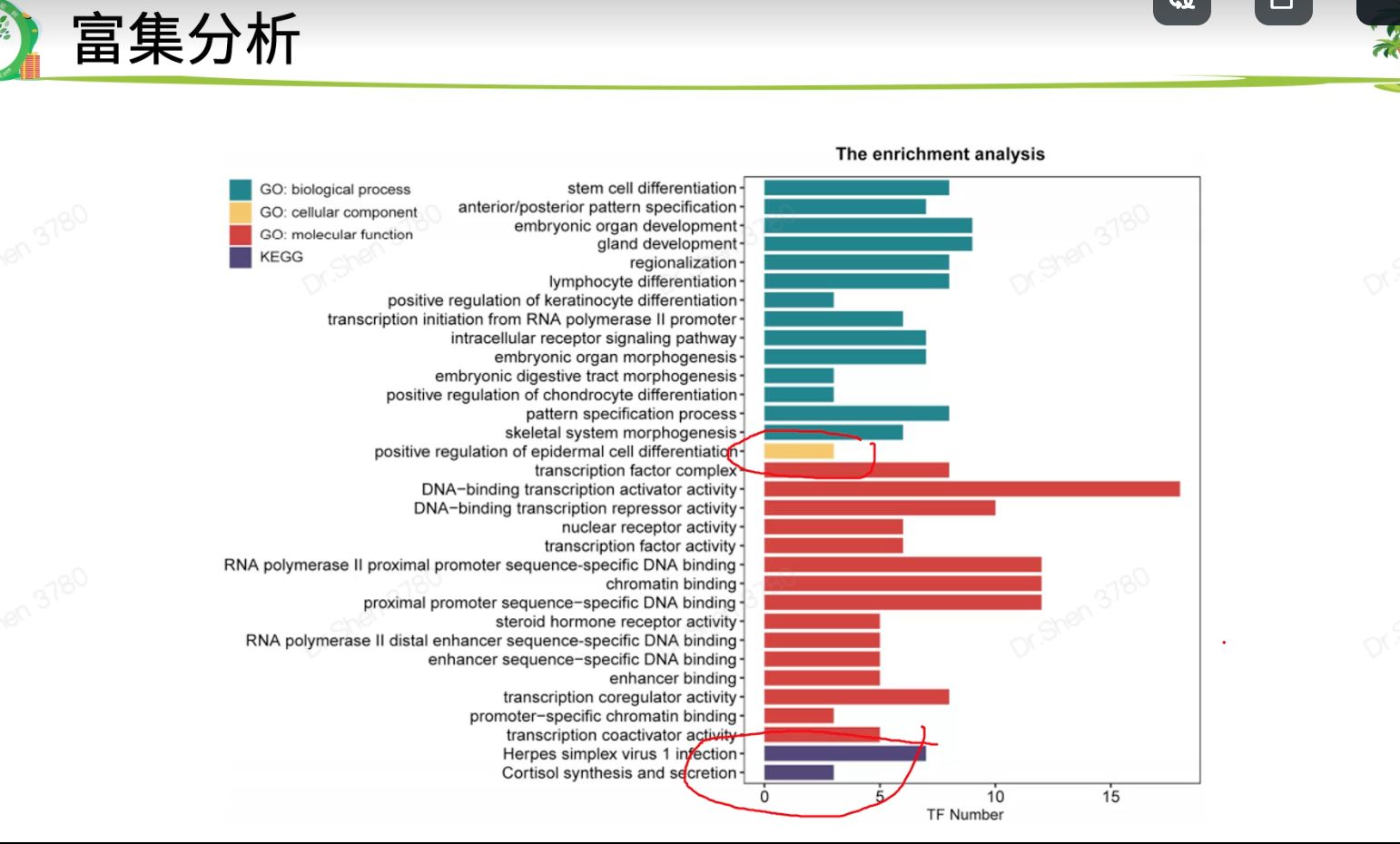
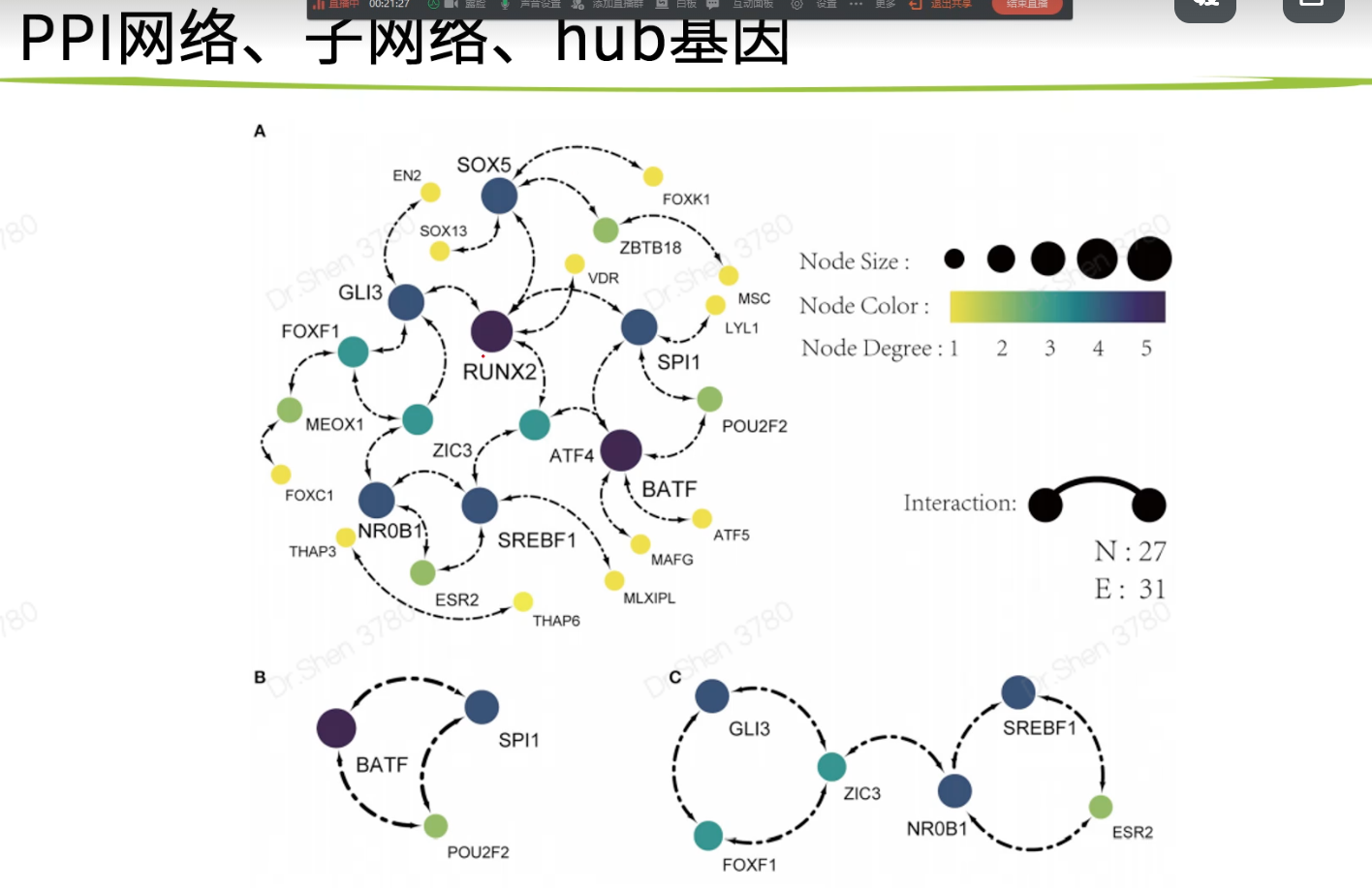
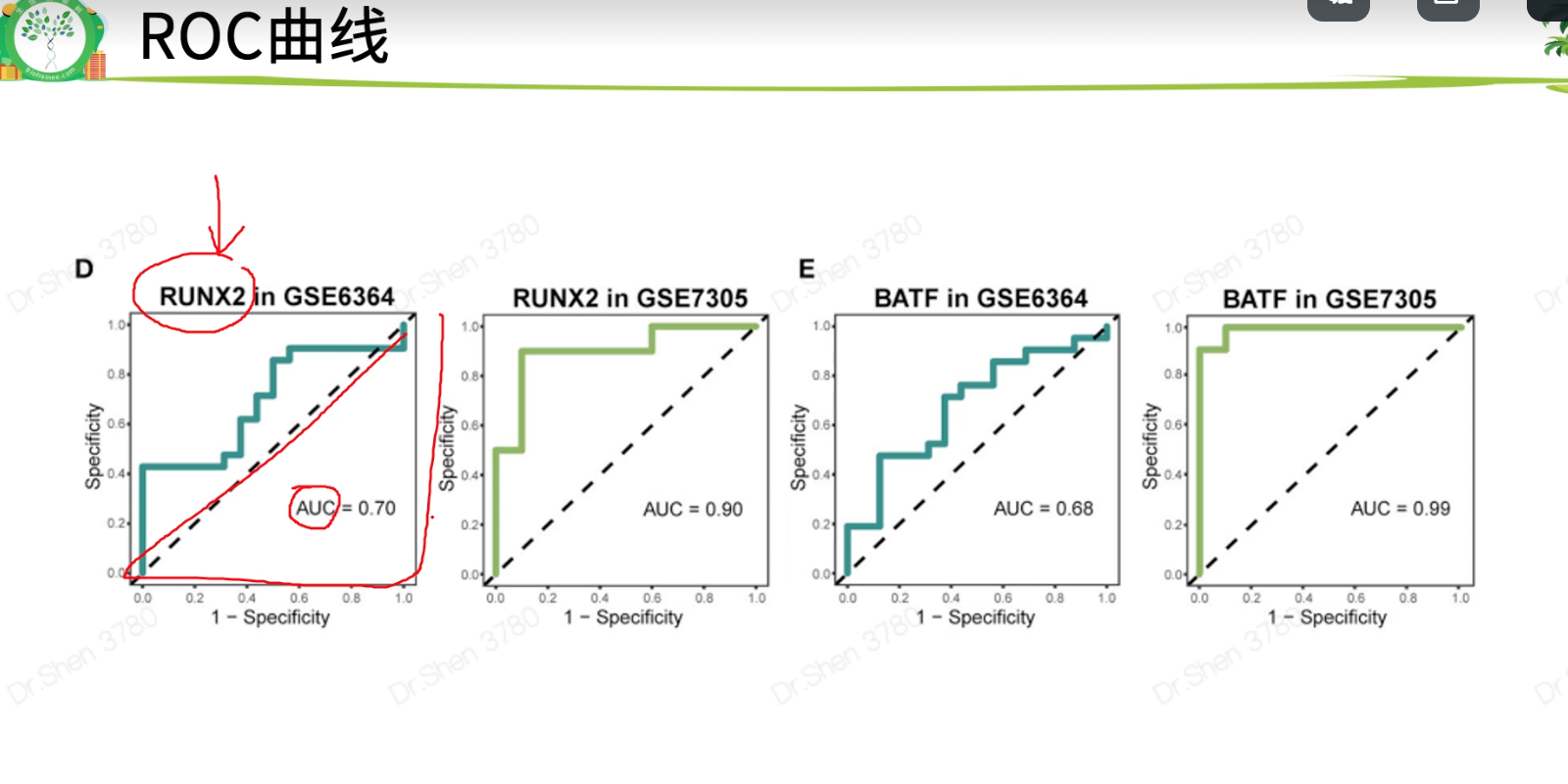
文献4 多数据合并

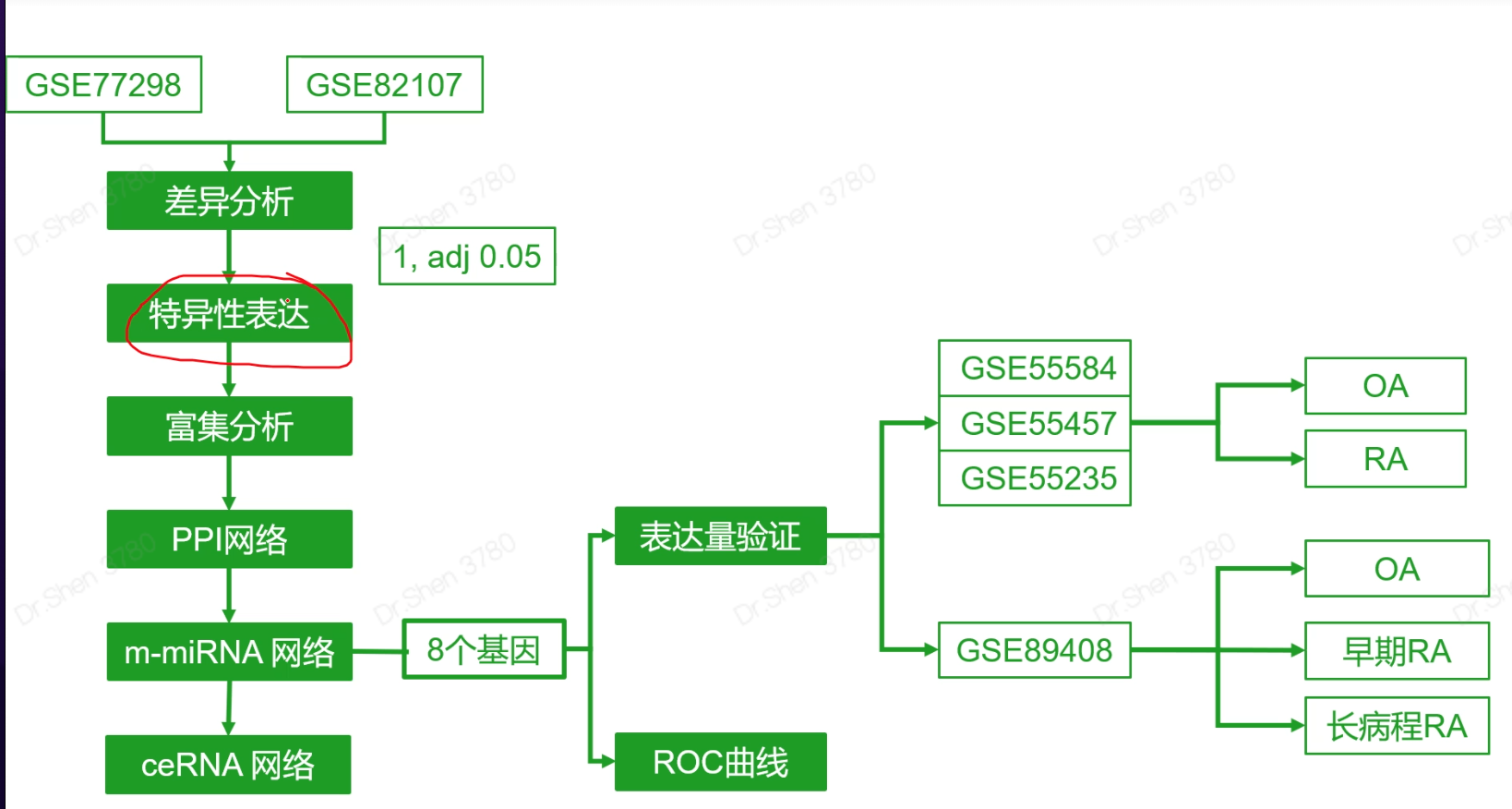
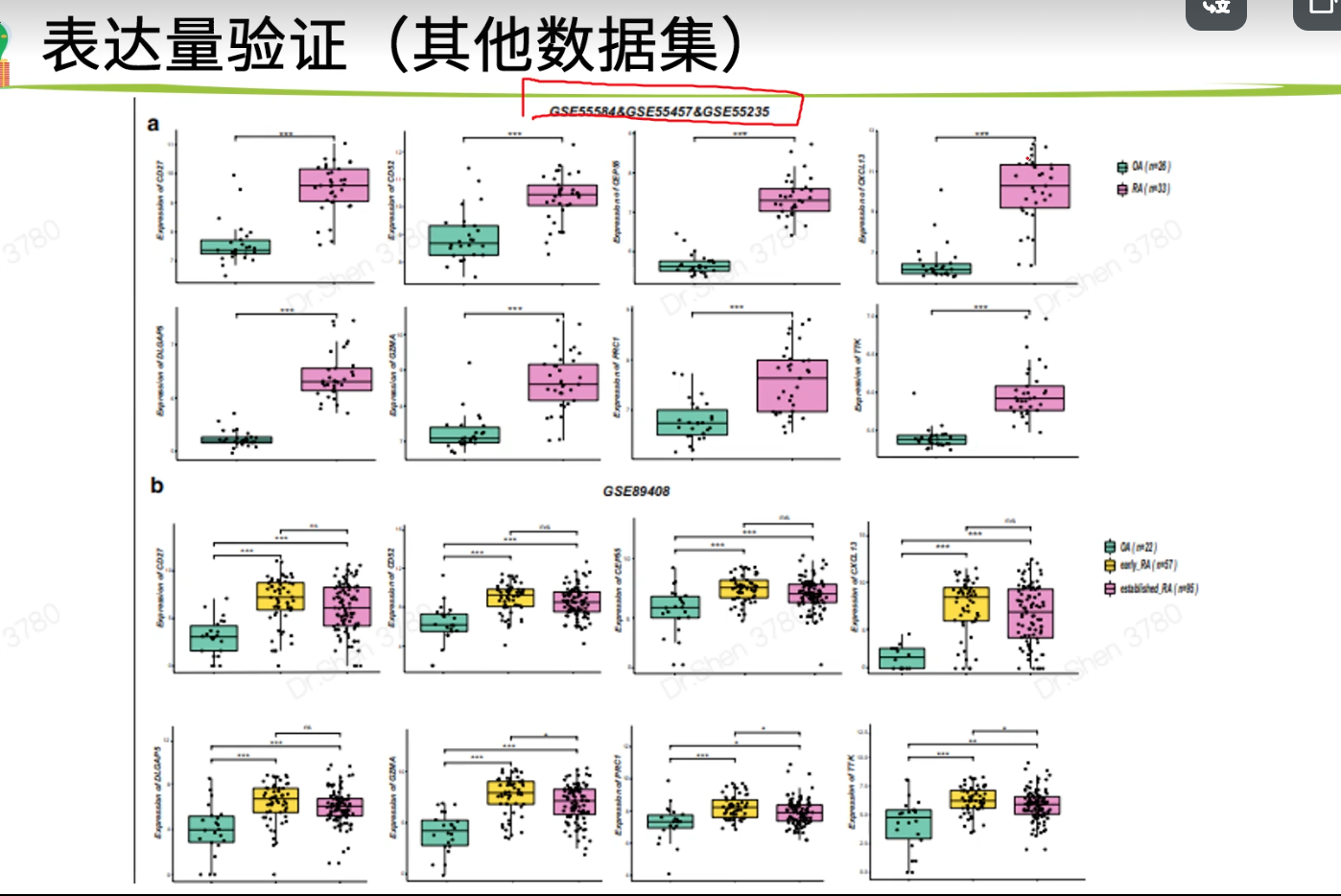
报错及分析