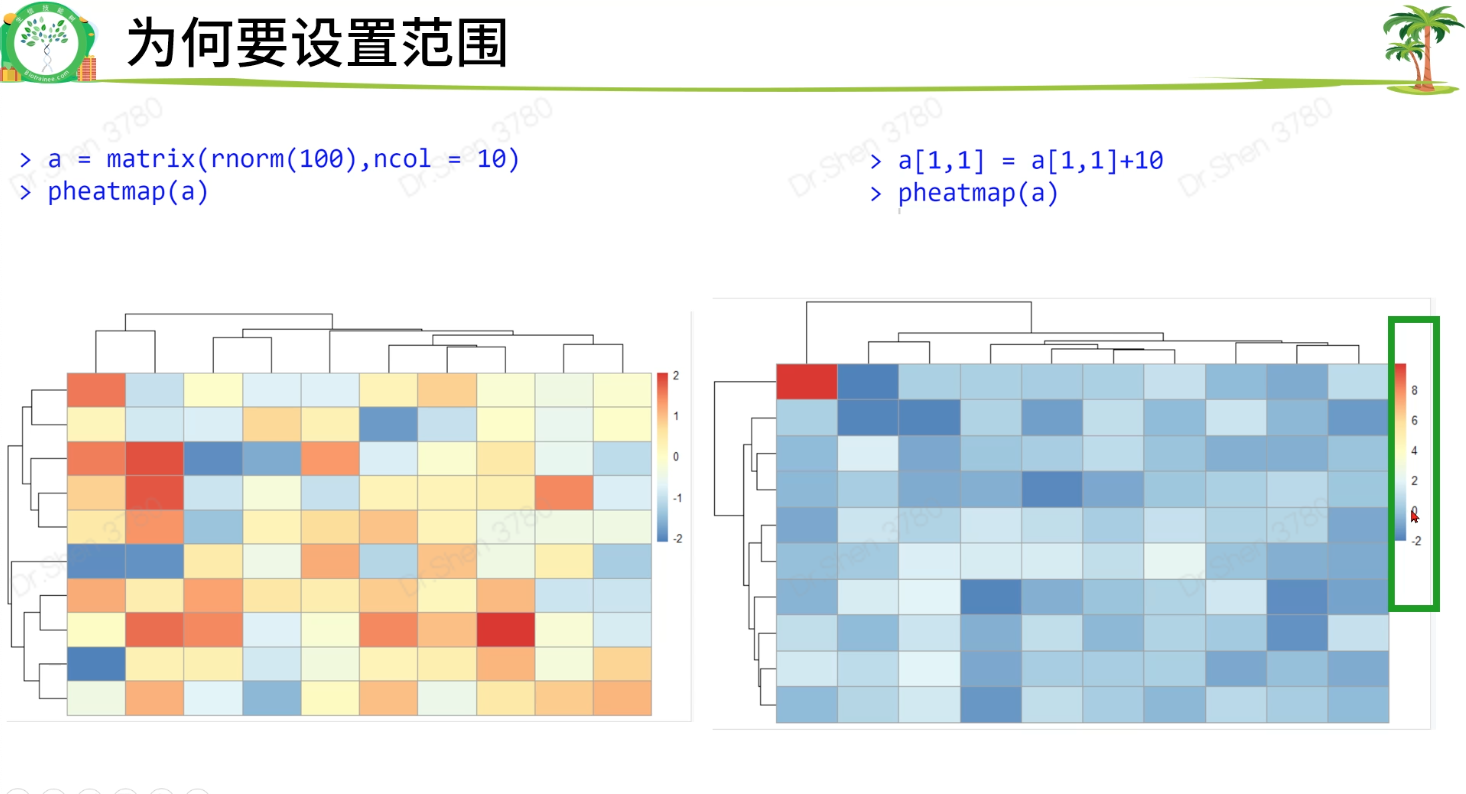
01、02是重点,需着重处理好,后续才能顺利,可形成模版
安装
options("repos"="https://mirrors.ustc.edu.cn/CRAN/")if(!require("BiocManager")) install.packages("BiocManager",update = F,ask = F)options(BioC_mirror="https://mirrors.ustc.edu.cn/bioc/")cran_packages <- c('tidyr','tibble','dplyr','stringr','ggplot2','ggpubr','factoextra','FactoMineR','devtools','cowplot','patchwork','basetheme','paletteer','AnnoProbe','ggthemes','VennDiagram','tinyarray')Biocductor_packages <- c('GEOquery','hgu133plus2.db','ggnewscale',"limma","impute","GSEABase","GSVA","clusterProfiler","org.Hs.eg.db","preprocessCore","enrichplot","ggplotify")for (pkg in cran_packages){if (! require(pkg,character.only=T) ) {install.packages(pkg,ask = F,update = F)require(pkg,character.only=T)}}for (pkg in Biocductor_packages){if (! require(pkg,character.only=T) ) {BiocManager::install(pkg,ask = F,update = F)require(pkg,character.only=T)}}#前面的所有提示和报错都先不要管。主要看这里for (pkg in c(Biocductor_packages,cran_packages)){require(pkg,character.only=T)}#没有任何提示就是成功了,如果有warningxx包不存在,用library检查一下。#library报错,就单独安装。library(hgu133plus2.db)
下载及检查数据
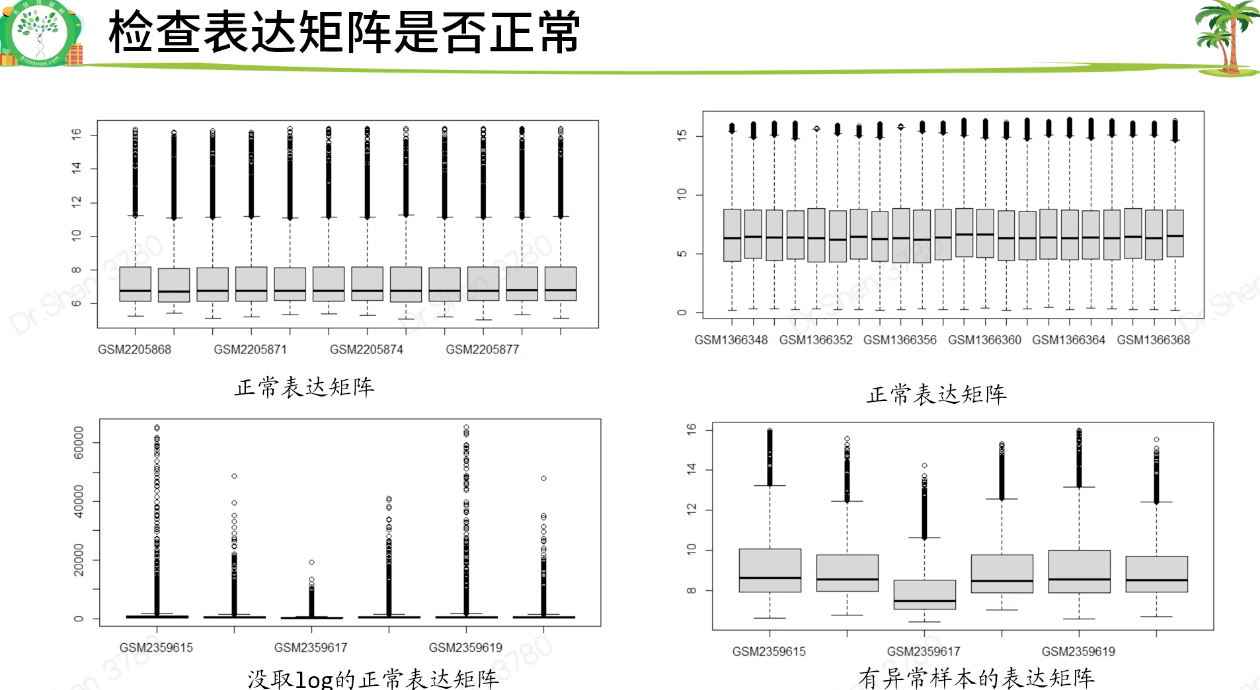
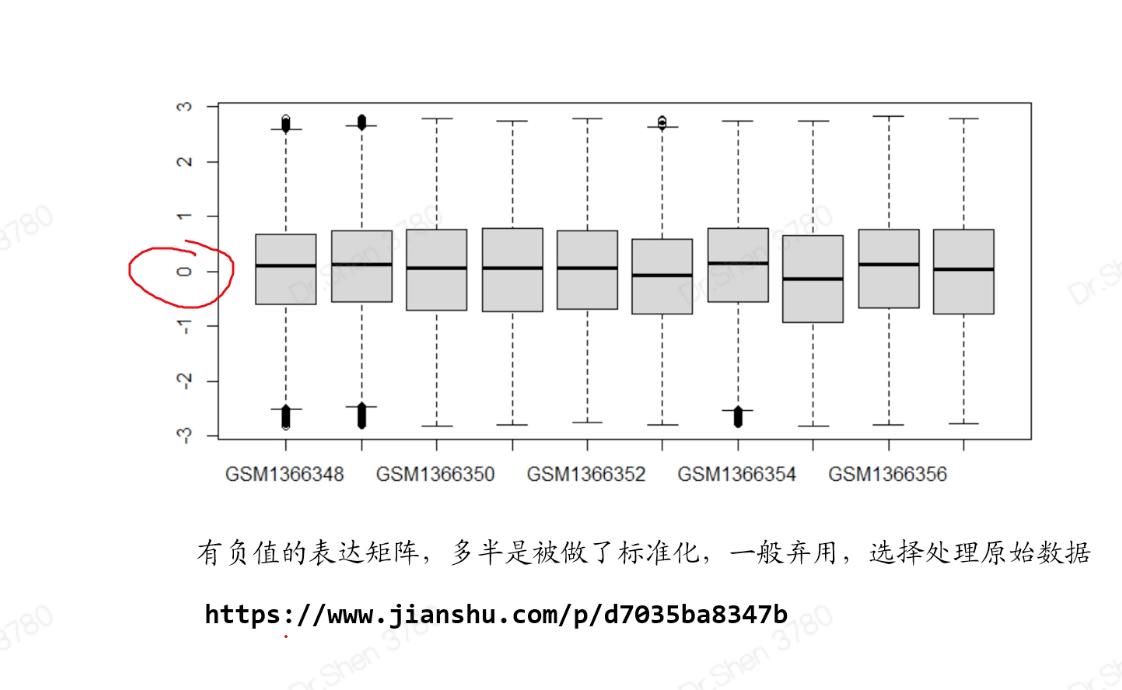
#切记先听课再跑代码#数据下载rm(list = ls())library(GEOquery)#先去网页确定是否是表达芯片数据,不是的话不能用本流程。#功能参数不需要更改#getGEO具有判断机制,如果本地有,则不会再下载gse_number = "GSE56649"eSet <- getGEO(gse_number, destdir = '.', getGPL = F)class(eSet)length(eSet)eSet = eSet[[1]]#(1)提取表达矩阵expexp <- exprs(eSet)dim(exp)exp[1:4,1:4]#列出前4行4列的情况#检查矩阵是否正常,如果是空的就会报错。对于空的、负值的、异常值的矩阵需要处理原始数据#如果表达矩阵为空,大多数是转录组数据,不能用这个流程(后面另讲)。#自行判断是否需要log。一般数值<20,则取过log;若是几百上千,则未取exp = log2(exp+1)boxplot(exp)#(2)提取临床信息pd <- pData(eSet)#(3)让exp列名与pd的行名顺序完全一致p = identical(rownames(pd),colnames(exp));pif(!p) exp = exp[,match(rownames(pd),colnames(exp))]#(4)提取芯片平台编号gpl_number <- eSet@annotation;gpl_numbersave(gse_number,pd,exp,gpl_number,file = "step1output.Rdata")
实验分组&探针注释
# Group(实验分组)和ids(探针注释)rm(list = ls())load(file = "step1output.Rdata")library(stringr)# 标准流程代码是二分组,多分组数据的分析后面另讲# 生成Group向量的三种常规方法,三选一,选谁就把第几个逻辑值写成T,另外两个为F。如果三种办法都不适用,可以继续往后写else ifif(F){# 1.Group----# 第一种方法,有现成的可以用来分组的列Group = pd$`disease state:ch1`}else if(F){# 第二种方法,自己生成Group = c(rep("RA",times=13),rep("control",times=9))Group = rep(c("RA","control"),times = c(13,9))}else if(T){# 第三种方法,使用字符串出理的函数获取分组Group=ifelse(str_detect(pd$source_name_ch1,"control"),"control","RA")}# 需要把Group转换成因子,并设置参考水平,指定levels,对照组在前,处理组在后Group = factor(Group,levels = c("control","RA"))Group
探针注释:探针与基因的对应关系

gpl文件解析
自主注释流程
可能存在一个探针对应多个基因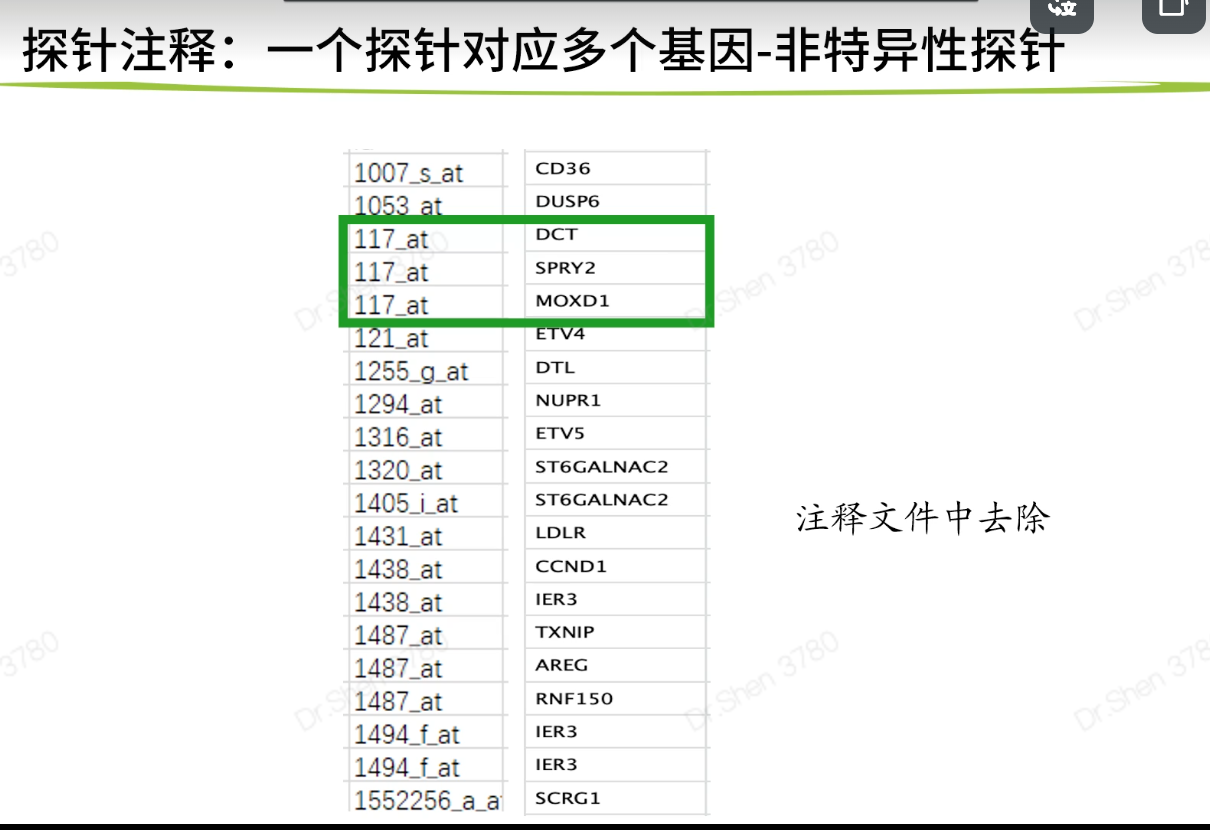
#2.探针注释的获取-----------------#捷径library(tinyarray)find_anno(gpl_number)ids <- AnnoProbe::idmap('GPL570')#四种方法,方法1里找不到就从方法2找,以此类推。#不同的方法,获取的数值可能有些差异,但被允许#方法1 BioconductorR包(最常用)gpl_number#http://www.bio-info-trainee.com/1399.htmlif(!require(hgu133plus2.db))BiocManager::install("hgu133plus2.db")library(hgu133plus2.db)ls("package:hgu133plus2.db")ids <- toTable(hgu133plus2SYMBOL)head(ids)# 方法2 读取GPL网页的表格文件,按列取子集##https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL570if(F){#注:表格读取参数、文件列名不统一,活学活用,有的表格里没有symbol列,也有的GPL平台没有提供注释表格b = read.table("GPL570-55999.txt",header = T,quote = "\"",sep = "\t",check.names = F)colnames(b)ids2 = b[,c("ID","Gene Symbol")] #按列名取子集colnames(ids2) = c("probe_id","symbol")ids2 = ids2[ids2$symbol!="" & !str_detect(ids2$symbol,"///"),]} #去除存在///的探针# 方法3 官网下载注释文件并读取##http://www.affymetrix.com/support/technical/byproduct.affx?product=hg-u133-plus# 方法4 自主注释#https://mp.weixin.qq.com/s/mrtjpN8yDKUdCSvSUuUwcAsave(exp,Group,ids,gse_number,file = "step2output.Rdata")
差异分析图
rm(list = ls())load(file = "step1output.Rdata")load(file = "step4output.Rdata")#1.火山图----library(dplyr)library(ggplot2)dat = deg[!duplicated(deg$symbol),]p <- ggplot(data = dat,aes(x = logFC,y = -log10(P.Value))) +geom_point(alpha=0.4, size=3.5,aes(color=change)) +ylab("-log10(Pvalue)")+scale_color_manual(values=c("blue", "grey","red"))+geom_vline(xintercept=c(-logFC_t,logFC_t),lty=4,col="black",lwd=0.8) +geom_hline(yintercept = -log10(P.Value_t),lty=4,col="black",lwd=0.8) +theme_bw()pfor_label <- dat%>%filter(symbol %in% c("HADHA","LRRFIP1")) #添加指定基因volcano_plot <- p +geom_point(size = 3, shape = 1, data = for_label) +ggrepel::geom_label_repel(aes(label = symbol),data = for_label,color="black") #对数据进行筛选,允许对不同图层进行特定设置volcano_plot#2.差异基因热图----load(file = 'step2output.Rdata')# 表达矩阵行名替换exp = exp[dat$probe_id,]rownames(exp) = dat$symbolif(F){#全部差异基因cg = dat$symbol[dat$change !="stable"]length(cg)}else{#取前10上调和前10下调library(dplyr)dat2 = dat %>%filter(change!="stable") %>%arrange(logFC)cg = c(head(dat2$symbol,10),tail(dat2$symbol,10))}n=exp[cg,]dim(n)#差异基因热图library(pheatmap)annotation_col=data.frame(group=Group)rownames(annotation_col)=colnames(n)heatmap_plot <- pheatmap(n,show_colnames =F,scale = "row",#cluster_cols = F,annotation_col=annotation_col,breaks = seq(-3,3,length.out = 100))heatmap_plot#拼图library(patchwork)library(ggplotify)volcano_plot +as.ggplot(heatmap_plot)# 3.感兴趣基因的相关性----library(corrplot)g = deg$symbol[1:10] # 换成自己感兴趣的基因gM = cor(t(exp[g,]))pheatmap(M)library(paletteer)my_color = rev(paletteer_d("RColorBrewer::RdYlBu"))my_color = colorRampPalette(my_color)(10)corrplot(M, type="upper",method="pie",order="hclust",col=my_color,tl.col="black",tl.srt=45)library(cowplot)cor_plot <- recordPlot()#抠图# 拼图load("pca_plot.Rdata")plot_grid(pca_plot,cor_plot,volcano_plot,heatmap_plot$gtable)dev.off()#保存pdf("deg.pdf", width = 12,height = 12 )#修改图的大小plot_grid(pca_plot,cor_plot,volcano_plot,heatmap_plot$gtable)dev.off()
主成分分析


rm(list = ls())load(file = "step1output.Rdata")load(file = "step2output.Rdata")#输入数据:exp和Group#Principal Component Analysis#http://www.sthda.com/english/articles/31-principal-component-methods-in-r-practical-guide/112-pca-principal-component-analysis-essentials# 1.PCA 图----dat=as.data.frame(t(exp))library(FactoMineR)library(factoextra)dat.pca <- PCA(dat, graph = FALSE)pca_plot <- fviz_pca_ind(dat.pca,geom.ind = "point", # show points only (nbut not "text")col.ind = Group, # color by groupspalette = c("#00AFBB", "#E7B800"),addEllipses = TRUE, # Concentration ellipseslegend.title = "Groups")pca_plotsave(pca_plot,file = "pca_plot.Rdata")# 2.top 1000 sd 热图----cg=names(tail(sort(apply(exp,1,sd)),1000))n=exp[cg,]# 直接画热图,对比不鲜明library(pheatmap)annotation_col=data.frame(group=Group) #指示条rownames(annotation_col)=colnames(n)pheatmap(n,show_colnames =F,show_rownames = F,annotation_col=annotation_col)# 按行标准化pheatmap(n,show_colnames =F,show_rownames = F,annotation_col=annotation_col,scale = "row", #实现标准化breaks = seq(-3,3,length.out = 100) #设置色带)dev.off()# 关于scale的进一步学习:zz.scale.R
DEG差异分析、可视化
rm(list = ls())load(file = "step2output.Rdata")#差异分析,用limma包来做#需要表达矩阵和Group,不需要改library(limma)design=model.matrix(~Group)fit=lmFit(exp,design)fit=eBayes(fit)deg=topTable(fit,coef=2,number = Inf)#为deg数据框添加几列#1.加probe_id列,把行名变成一列library(dplyr)deg <- mutate(deg,probe_id=rownames(deg))#2.加上探针注释ids = ids[!duplicated(ids$symbol),] #随机去重#其他去重方式在zz.去重方式.Rdeg <- inner_join(deg,ids,by="probe_id")nrow(deg)#3.加change列,标记上下调基因logFC_t=1P.Value_t = 0.05k1 = (deg$P.Value < P.Value_t)&(deg$logFC < -logFC_t)k2 = (deg$P.Value < P.Value_t)&(deg$logFC > logFC_t)deg <- mutate(deg,change = ifelse(k1,"down",ifelse(k2,"up","stable")))table(deg$change)#4.加ENTREZID列,用于富集分析(symbol转entrezid,然后inner_join)library(clusterProfiler)library(org.Hs.eg.db)s2e <- bitr(deg$symbol,fromType = "SYMBOL",toType = "ENTREZID",OrgDb = org.Hs.eg.db)#人类#其他物种http://bioconductor.org/packages/release/BiocViews.html#___OrgDbdeg <- inner_join(deg,s2e,by=c("symbol"="SYMBOL"))save(Group,deg,logFC_t,P.Value_t,gse_number,file = "step4output.Rdata")
富集分析(GSEA)
rm(list = ls())load(file = 'step4output.Rdata')library(clusterProfiler)library(ggthemes)library(org.Hs.eg.db)library(dplyr)library(ggplot2)library(stringr)library(enrichplot)# 1.GO 富集分析----#(1)输入数据gene_up = deg$ENTREZID[deg$change == 'up']gene_down = deg$ENTREZID[deg$change == 'down']gene_diff = c(gene_up,gene_down)#(2)富集#以下步骤耗时很长,设置了存在即跳过if(!file.exists(paste0(gse_number,"_GO.Rdata"))){ego <- enrichGO(gene = gene_diff,OrgDb= org.Hs.eg.db,ont = "ALL",readable = TRUE)ego_BP <- enrichGO(gene = gene_diff,OrgDb= org.Hs.eg.db,ont = "BP",readable = TRUE)#ont参数:One of "BP", "MF", and "CC" subontologies, or "ALL" for all three.save(ego,ego_BP,file = paste0(gse_number,"_GO.Rdata"))}load(paste0(gse_number,"_GO.Rdata"))#(3)可视化#条带图barplot(ego)barplot(ego, split = "ONTOLOGY", font.size = 10,showCategory = 5) +facet_grid(ONTOLOGY ~ ., space = "free_y",scales = "free_y")#气泡图dotplot(ego)dotplot(ego, split = "ONTOLOGY", font.size = 10,showCategory = 5) +facet_grid(ONTOLOGY ~ ., space = "free_y",scales = "free_y")#(3)展示top通路的共同基因,要放大看。#gl 用于设置下图的颜色gl = deg$logFCnames(gl)=deg$ENTREZID#Gene-Concept Networkcnetplot(ego,categorySize="pvalue", foldChange=gl,colorEdge = TRUE,showCategory = 3)cnetplot(ego, showCategory = 3,foldChange=gl, circular = TRUE, colorEdge = TRUE)# 2.KEGG pathway analysis----#上调、下调、差异、所有基因#(1)输入数据gene_up = deg[deg$change == 'up','ENTREZID']gene_down = deg[deg$change == 'down','ENTREZID']gene_diff = c(gene_up,gene_down)#(2)对上调/下调/所有差异基因进行富集分析if(!file.exists(paste0(gse_number,"_KEGG.Rdata"))){kk.up <- enrichKEGG(gene = gene_up,organism = 'hsa')kk.down <- enrichKEGG(gene = gene_down,organism = 'hsa')kk.diff <- enrichKEGG(gene = gene_diff,organism = 'hsa')save(kk.diff,kk.down,kk.up,file = paste0(gse_number,"_KEGG.Rdata"))}load(paste0(gse_number,"_KEGG.Rdata"))#(3)看看富集到了吗?https://mp.weixin.qq.com/s/NglawJgVgrMJ0QfD-YRBQgtable(kk.diff@result$p.adjust<0.05)table(kk.up@result$p.adjust<0.05)table(kk.down@result$p.adjust<0.05)#(4)双向图# 富集分析所有图表默认都是用p.adjust,富集不到可以退而求其次用p值,在文中说明即可down_kegg <- kk.down@result %>%filter(pvalue<0.05) %>% #筛选行mutate(group=-1) #新增列up_kegg <- kk.up@result %>%filter(pvalue<0.05) %>%mutate(group=1)source("kegg_plot_function.R")g_kegg <- kegg_plot(up_kegg,down_kegg)g_kegg#g_kegg +scale_y_continuous(labels = c(2,0,2,4,6))# 3.GSEA作kegg和GO富集分析----#https://www.yuque.com/xiaojiewanglezenmofenshen/dbwkg1/ytawgg#(1)查看示例数据data(geneList, package="DOSE")#(2)将我们的数据转换成示例数据的格式geneList=deg$logFCnames(geneList)=deg$ENTREZIDgeneList=sort(geneList,decreasing = T)#(3)GSEA富集kk_gse <- gseKEGG(geneList = geneList,organism = 'hsa',verbose = FALSE)down_kegg<-kk_gse[kk_gse$pvalue<0.05 & kk_gse$enrichmentScore < 0,];down_kegg$group=-1up_kegg<-kk_gse[kk_gse$pvalue<0.05 & kk_gse$enrichmentScore > 0,];up_kegg$group=1#(4)可视化g2 = kegg_plot(up_kegg,down_kegg)g2# 4.能看懂的资料越来越多----# GSEA学习更多:https://www.yuque.com/docs/share/a67a180f-dd2b-4f6f-96c2-68a4b86fe862?## 富集分析学习更多:http://yulab-smu.top/clusterProfiler-book/index.html# 弦图:https://www.yuque.com/xiaojiewanglezenmofenshen/dbwkg1/dgs65p# GOplot:https://mp.weixin.qq.com/s/LonwdDhDn8iFUfxqSJ2Wew# 网上的资料和宝藏无穷无尽,学好R语言慢慢发掘~
if(F){a = 1 #限速步骤save(a,file = "a.Rdata")}load("a.Rdata")if(!file.exists("a.Rdata")){a = 1 #限速步骤save(a,file = "a.Rdata")}load("a.Rdata")
问题和解答