load("TCGA-CHOL_DEG.Rdata")load("TCGA-CHOL.Rdata")cg2 = rownames(DEG2)[DEG2$change !="NOT"]library(tinyarray)library(edgeR)dat = log2(cpm(exp)+1)draw_heatmap(dat[cg2,],Group,cluster_cols = F)dat2Group2 = sort(Group)dat2 = dat[,order(Group)]draw_heatmap(dat2[cg2,],Group2,cluster_cols = F)
三大R包差异分析
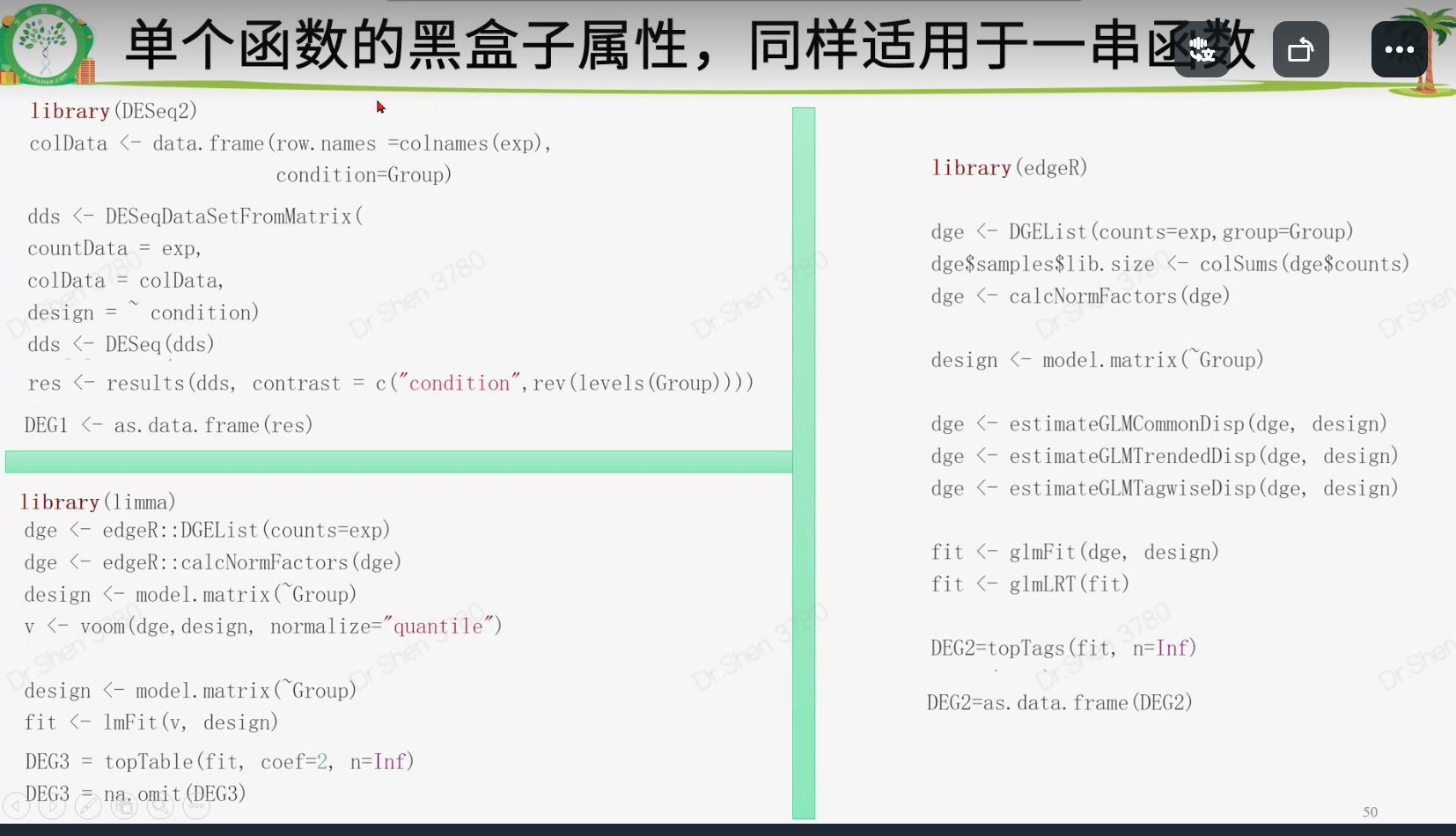
rm(list = ls())load("DHA.Rdata")table(Group)#> Group#> DMSO DHA#> 3 3#deseq2----library(DESeq2)colData <- data.frame(row.names =colnames(exp),condition=Group)if(!file.exists(paste0(proj,"_dd.Rdata"))){dds <- DESeqDataSetFromMatrix(countData = exp,colData = colData,design = ~ condition)dds <- DESeq(dds)save(dds,file = paste0(proj,"_dd.Rdata"))}load(file = paste0(proj,"_dd.Rdata"))class(dds)#> [1] "DESeqDataSet"#> attr(,"package")#> [1] "DESeq2"res <- results(dds, contrast = c("condition",rev(levels(Group))))#constrastc("condition",rev(levels(Group)))#> [1] "condition" "DHA" "DMSO"class(res)#> [1] "DESeqResults"#> attr(,"package")#> [1] "DESeq2"DEG1 <- as.data.frame(res)DEG1 <- DEG1[order(DEG1$pvalue),]DEG1 = na.omit(DEG1)head(DEG1)#> baseMean log2FoldChange lfcSE stat pvalue padj#> CDH6 5006.549 -4.030551 0.10671818 -37.76818 0.000000e+00 0.000000e+00#> CXCL8 22087.170 4.958524 0.12501648 39.66296 0.000000e+00 0.000000e+00#> HMOX1 46652.918 2.995161 0.08194416 36.55124 1.704000e-292 1.163094e-288#> THBD 2832.828 5.025714 0.13858308 36.26499 5.768388e-288 2.952982e-284#> LIF 4105.206 4.812167 0.14857784 32.38819 4.025761e-230 1.648710e-226#> DUSP5 2620.024 4.334570 0.13469580 32.18044 3.314657e-227 1.131237e-223#添加change列标记基因上调下调logFC_t = 2pvalue_t = 0.05k1 = (DEG1$pvalue < pvalue_t)&(DEG1$log2FoldChange < -logFC_t);table(k1)#> k1#> FALSE TRUE#> 19726 751k2 = (DEG1$pvalue < pvalue_t)&(DEG1$log2FoldChange > logFC_t);table(k2)#> k2#> FALSE TRUE#> 19858 619DEG1$change = ifelse(k1,"DOWN",ifelse(k2,"UP","NOT"))table(DEG1$change)#>#> DOWN NOT UP#> 751 19107 619head(DEG1)#> baseMean log2FoldChange lfcSE stat pvalue padj#> CDH6 5006.549 -4.030551 0.10671818 -37.76818 0.000000e+00 0.000000e+00#> CXCL8 22087.170 4.958524 0.12501648 39.66296 0.000000e+00 0.000000e+00#> HMOX1 46652.918 2.995161 0.08194416 36.55124 1.704000e-292 1.163094e-288#> THBD 2832.828 5.025714 0.13858308 36.26499 5.768388e-288 2.952982e-284#> LIF 4105.206 4.812167 0.14857784 32.38819 4.025761e-230 1.648710e-226#> DUSP5 2620.024 4.334570 0.13469580 32.18044 3.314657e-227 1.131237e-223#> change#> CDH6 DOWN#> CXCL8 UP#> HMOX1 UP#> THBD UP#> LIF UP#> DUSP5 UP#edgeR----library(edgeR)dge <- DGEList(counts=exp,group=Group)dge$samples$lib.size <- colSums(dge$counts)dge <- calcNormFactors(dge)design <- model.matrix(~Group)dge <- estimateGLMCommonDisp(dge, design)dge <- estimateGLMTrendedDisp(dge, design)dge <- estimateGLMTagwiseDisp(dge, design)fit <- glmFit(dge, design)fit <- glmLRT(fit)DEG2=topTags(fit, n=Inf)class(DEG2)#> [1] "TopTags"#> attr(,"package")#> [1] "edgeR"DEG2=as.data.frame(DEG2)head(DEG2)#> logFC logCPM LR PValue FDR#> LIF 4.780301 7.069581 571.9517 2.112394e-126 4.695851e-122#> PTGS2 5.970830 7.152826 565.5336 5.258853e-125 5.845215e-121#> THBD 4.992993 6.534219 564.2212 1.014763e-124 7.519395e-121#> CDH6 -4.062412 7.384277 532.6938 7.326559e-118 4.071735e-114#> CXCL8 4.926708 9.496259 470.0676 3.097901e-104 1.377327e-100#> TMEM158 5.147555 6.279693 437.4408 3.903283e-97 1.446166e-93k1 = (DEG2$PValue < pvalue_t)&(DEG2$logFC < -logFC_t)k2 = (DEG2$PValue < pvalue_t)&(DEG2$logFC > logFC_t)DEG2$change = ifelse(k1,"DOWN",ifelse(k2,"UP","NOT"))head(DEG2)#> logFC logCPM LR PValue FDR change#> LIF 4.780301 7.069581 571.9517 2.112394e-126 4.695851e-122 UP#> PTGS2 5.970830 7.152826 565.5336 5.258853e-125 5.845215e-121 UP#> THBD 4.992993 6.534219 564.2212 1.014763e-124 7.519395e-121 UP#> CDH6 -4.062412 7.384277 532.6938 7.326559e-118 4.071735e-114 DOWN#> CXCL8 4.926708 9.496259 470.0676 3.097901e-104 1.377327e-100 UP#> TMEM158 5.147555 6.279693 437.4408 3.903283e-97 1.446166e-93 UPtable(DEG2$change)#>#> DOWN NOT UP#> 936 20586 708###limma----library(limma)dge <- edgeR::DGEList(counts=exp)dge <- edgeR::calcNormFactors(dge)design <- model.matrix(~Group)v <- voom(dge,design, normalize="quantile")fit <- lmFit(v, design)fit= eBayes(fit)DEG3 = topTable(fit, coef=2, n=Inf)DEG3 = na.omit(DEG3)k1 = (DEG3$P.Value < pvalue_t)&(DEG3$logFC < -logFC_t)k2 = (DEG3$P.Value < pvalue_t)&(DEG3$logFC > logFC_t)DEG3$change = ifelse(k1,"DOWN",ifelse(k2,"UP","NOT"))table(DEG3$change)#>#> DOWN NOT UP#> 846 20611 773head(DEG3)#> logFC AveExpr t P.Value adj.P.Val B change#> HMOX1 3.023387 9.933877 59.51263 1.613427e-10 1.895736e-06 15.06135 UP#> CXCL8 5.010797 8.042577 58.11745 1.896200e-10 1.895736e-06 14.56756 UP#> PLIN2 3.029546 9.754344 54.12555 3.077709e-10 1.895736e-06 14.47996 UP#> SQSTM1 2.528669 10.930792 51.44786 4.346774e-10 1.895736e-06 14.22328 UP#> AKR1C1 3.492536 8.480250 50.22935 5.116695e-10 1.895736e-06 13.91592 UP#> CDH6 -4.028740 6.272248 -52.96068 3.568837e-10 1.895736e-06 13.67540 DOWNtj = data.frame(deseq2 = as.integer(table(DEG1$change)),edgeR = as.integer(table(DEG2$change)),limma_voom = as.integer(table(DEG3$change)),row.names = c("down","not","up"));tj#> deseq2 edgeR limma_voom#> down 751 936 846#> not 19107 20586 20611#> up 619 708 773save(DEG1,DEG2,DEG3,Group,tj,file = paste0(proj,"_DEG.Rdata"))
可视化
library(ggplot2)library(tinyarray)exp[1:4,1:4]#> DMSO1 DMSO2 DMSO3 S25uMDNHA1#> MADD 1097 1586 1055 1527#> MAST2 2765 2951 2670 5872#> TRBJ2-5 2 1 1 1#> TRBJ2-4 1 1 1 1# cpm 去除文库大小的影响dat = log2(cpm(exp)+1)pca.plot = draw_pca(dat,Group);pca.plot
save(pca.plot,file = paste0(proj,"_pcaplot.Rdata"))cg1 = rownames(DEG1)[DEG1$change !="NOT"]cg2 = rownames(DEG2)[DEG2$change !="NOT"]cg3 = rownames(DEG3)[DEG3$change !="NOT"]h1 = draw_heatmap(dat[cg1,],Group)h2 = draw_heatmap(dat[cg2,],Group)h3 = draw_heatmap(dat[cg3,],Group)v1 = draw_volcano(DEG1,pkg = 1,logFC_cutoff = logFC_t)v2 = draw_volcano(DEG2,pkg = 2,logFC_cutoff = logFC_t)v3 = draw_volcano(DEG3,pkg = 3,logFC_cutoff = logFC_t)library(patchwork)(h1 + h2 + h3) / (v1 + v2 + v3) +plot_layout(guides = 'collect') &theme(legend.position = "none")
ggsave(paste0(proj,"_heat_vo.png"),width = 15,height = 10)
三大R包差异基因对比
UP=function(df){rownames(df)[df$change=="UP"]}DOWN=function(df){rownames(df)[df$change=="DOWN"]}up = intersect(intersect(UP(DEG1),UP(DEG2)),UP(DEG3))down = intersect(intersect(DOWN(DEG1),DOWN(DEG2)),DOWN(DEG3))dat = log2(cpm(exp)+1)hp = draw_heatmap(dat[c(up,down),],Group)#上调、下调基因分别画维恩图up_genes = list(Deseq2 = UP(DEG1),edgeR = UP(DEG2),limma = UP(DEG3))down_genes = list(Deseq2 = DOWN(DEG1),edgeR = DOWN(DEG2),limma = DOWN(DEG3))up.plot <- draw_venn(up_genes,"UPgene")down.plot <- draw_venn(down_genes,"DOWNgene")#维恩图拼图,终于搞定library(patchwork)#up.plot + down.plot# 拼图pca.plot + hp+up.plot +down.plot+ plot_layout(guides = "collect")ggsave(paste0(proj,"_heat_ve_pca.png"),width = 15,height = 10)
分组聚类的热图
library(ComplexHeatmap)library(circlize)col_fun = colorRamp2(c(-2, 0, 2), c("#2fa1dd", "white", "#f87669"))top_annotation = HeatmapAnnotation(cluster = anno_block(gp = gpar(fill = c("#f87669","#2fa1dd")),labels = levels(Group),labels_gp = gpar(col = "white", fontsize = 12)))m = Heatmap(t(scale(t(exp[c(up,down),]))),name = " ",col = col_fun,top_annotation = top_annotation,column_split = Group,show_heatmap_legend = T,border = F,show_column_names = F,show_row_names = F,use_raster = F,cluster_column_slices = F,column_title = NULL)m

