1. Download and install conda and other software
See tutorial: https://www.jianshu.com/p/a84cd44bac67
1.1 Conda installation
# image of Tsinghua university open-source website to download wget - chttps://mirrors.tuna.tsinghua.edu.cn/anaconda/miniconda/Miniconda2-latest-Linux-x86_64.sh
# Install bash Miniconda2- Latest-Linux-x86_64.sh# change the image source configuration conda config - add channels athttps://mirrors.tuna.tsinghua.edu.cn/anaconda/pkgs/free
conda config --add channelshttps://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/conda-forgeconda config --add channelshttps://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/biocondaconda config --set show_channel_urls yes
1.2 Create a small environment
Create a software installation environment called Ymy
conda cret-n ymy Python =3View the current Conda environmentconda info -- envsActivate Conda’s RNA environmentconda activate RNAExit the small environmentconda deactivate ymy
1.3 Software installation
Install Fastp conda Install -c bioconda Fastp
Install trim_galore conda install -y trim_galore
2. Fastp quality control:
2.1. Single two-terminal data processing:
fastp -i in.R1.fq.gz -I in.R2.fq.gz -o out.R1.fq.gz -O out.R2.fq.gz
2.2. Batch processing:
ls *_1.fastq | while read id;do "fastp -i $id -o ${id%_*}.1.fastp.fq -I ${id%_*}_2.fastq -O ${id%_*}_2.fastp.fq -h ${id%_*}.html ";done
2.3. Html&json format report
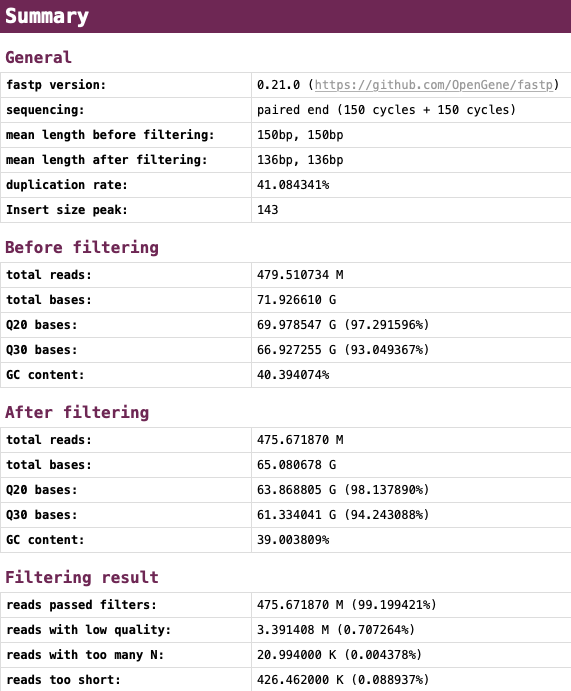
Filtering results: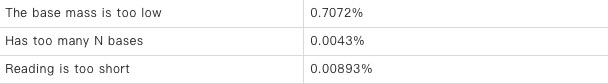
3.Trim_galore Quality Control Process
3.1. Single two-terminal data processing:
trim_galore /... /in.R1.fastq /... / In.r2.fastQ-Q 25 --length 50-e 0.1 -- Stringency 5-o /... / -a adapter1 -a2 adapter2 --paired
3.2. Batch processing of double-ended data:
cat trim.sh#! /bin/bashDir =/public/home/yuanmy/q1(be sure to use absolute path here)cat config |while read iddoarr=${id}fq1=${arr[0]}fq2=${arr[1]}Nohup trim_galore-q 20 --phred33 --length 36-e 0.1 -- Stringency 3 -- Paired -o $dir $fq1 $fQ2 &done
3.3. Interpretation of results:
Generate three intermediate files:
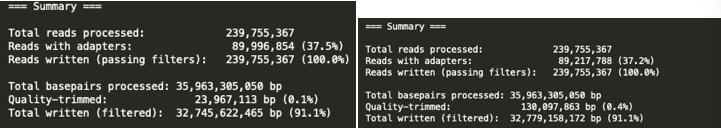
The number of bases trimmed by TRIM Galore was smaller than that of fastp:
| Trim Method | Trimed bases |
|---|---|
| Trim filtered base | 154064976 bp |
| Fastp filtered base | 7178479 bp |

