01. Find the virus:
ssh yuanmy@10.71.115.199cd /public/home/shared_lab/first11/Fasta_file/ls-lh# to see the data size, there are 11 pairs of PE data
02.Single-ended reads
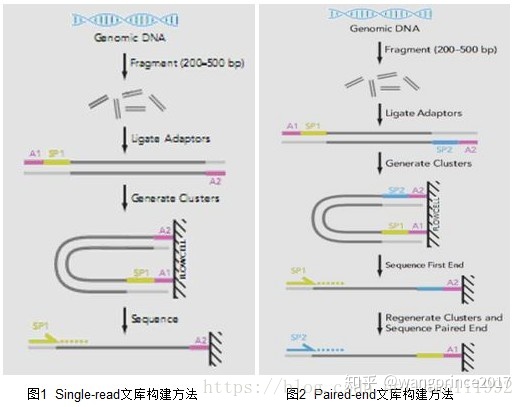
Firstly, single-read fragment DNA samples to 200-500bp fragments. The primer sequence is complementary to one end of the DNA fragment, and then an adaptor is added at the end to fix the fragment on the flow cell to generate the DNA cluster, and the sequence was sequenced by machine and read at the single end.
03.Paried-ended reads
Double-end sequencing (Paired-end) refers to adding sequencing primer binding sites on both ends of the terminals when constructing the DNA library to be tested. After the first round of sequencing is completed, the template chain of the first round of sequencing is removed. Paired sequencing module was used to guide the regeneration and amplification of the Paired chain at its original location.
To reach the number of templates used in the second round of sequencing,
Summary: “single” and “double” are targeted for the introduction of sequencing primers that are 1 or 2
As shown in figure: