前面我们通过 GATK 的 mutect 流程找到了 somatic mutations,但是只有突变的染色体和坐标信息,我们并不知道该突变发生在哪个基因或者哪号外显子,显然通过坐标进行手动查看的方法是不科学的。应该通过软件来实现,这里我们比较四个vcf注释软件的安装及使用:vep、annovar、gatk funcatator、snpeff
这一节我们将比较不同注释软件(vep,annovar,gatk funcatator,snpeff) 的安装及使用,按照我的代码习惯,都需要一个 config3 文件来完成批量处理,与前面的 config 文件区别在于,这里仅剩下肿瘤的样本名:
$ cat configcase1_biorep_A_techrepcase2_biorep_Acase3_biorep_Acase4_biorep_Acase5_biorep_A......
ANNOVAR
安装
cd ~/wes_cancer/biosoft# wget 下载地址tar -zxvf annovar.latest.tar.gzcd annovarnohup ./annotate_variation.pl -downdb -webfrom annovar gnomad_genome --buildver hg38 humandb/ >down.log 2>&1 &
数据量非常大,不过看个人所需下载,我这里其实下载了很多不必要的注释文件,当然可能以后会用到:
41G humandb/
如果想下载指定数据库的注释文件,可以先获取数据库注释文件列表
$ ./annotate_variation.pl -downdb -buildver hg38 -webfrom annovar avdblist hg38_list/$ cat hg38_list/hg38_avdblist.txthg38_abraom.txt.gz 20191205 23148851hg38_abraom.txt.idx.gz 20191205 9831917hg38_avsnp142.txt.gz 20191205 1282852569hg38_avsnp142.txt.idx.gz 20191205 212764014hg38_avsnp144.txt.gz 20191205 1671400214hg38_avsnp144.txt.idx.gz 20191205 215204030hg38_avsnp147.txt.gz 20191205 1775686247hg38_avsnp147.txt.idx.gz 20191205 222202148hg38_avsnp150.txt.gz 20191205 3794169304hg38_avsnp150.txt.idx.gz 20191205 234261780......
注释
在曾老师的博客中有多篇博文介绍它的用法,但主要看:ANNOVAR软件用法还可以更复杂:http://www.bio-info-trainee.com/4007.html,主要有3种类型的注释:
- 基于基因的注释,
exonic,splicing,ncRNA,UTR5,UTR3,intronic,upstream,downstream,intergenic,使用geneanno子命令。 - 基于区域的注释,
cytoBand,TFBS,SV,bed,GWAS,ENCODE,enhancers,repressors,promoters,使用regionanno子命令。只考虑位点坐标 - 基于数据库的过滤,
dbSNP,ExAC,ESP6500,cosmic,gnomad,1000genomes,clinvar使用filter子命令。 考虑位点坐标同时关心突变碱基情况。
而本文中,用 ANNOVAR 对 vcf 文件所做的是基于基因的注释,注释脚本annovar.sh如下:
cat config | while read iddoecho "start ANNOVAR for ${id} " `date`~/biosoft/annovar/table_annovar.pl ./6.mutect/${id}_filter.vcf ~/biosoft/annovar/humandb/ \-buildver hg38 \-out ./7.annotation/annovar/${id} \-remove \-protocol refGene,knownGene,clinvar_20170905 \-operation g,g,f \-nastring . \-vcfinputecho "end ANNOVAR for ${id} " `date`done
注释的速度非常快,一个样本大概就是6~7秒就可以注释完,每个样本输出3个文件:
case1_biorep_A_techrep.avinputcase1_biorep_A_techrep.hg38_multianno.txtcase1_biorep_A_techrep.hg38_multianno.vcf
在文件case1_biorep_A_techrep.hg38_multianno.txt中的一个突变记录位点注释上的信息如下:
chr1 6146376 6146376 G T exonic CHD5 . nonsynonymous SNV CHD5:NM_015557:exon11:c.C1638A:p.N546K exonic CHD5 . nonsynonymous SNV CHD5:uc001amb.3:exon11:c.C1638A:p.N546K,CHD5:uc057btb.1:exon11:c.C1638A:p.N546K . . . . . 0.25 .75 chr1 6146376 . G T . PASS DP=184;ECNT=1;NLOD=22.56;N_ART_LOD=-1.888e+00;POP_AF=1.000e-05;P_CONTAM=0.00;P_GERMLINE=-4.514e+01;TLOD=5.41 GT:AD:AF:F1R2:F2R1:MBQ:MFRL:MMQ:MPOS:SA_MAP_AF:SA_POST_PROB 0/1:97,3:0.058:48,1:49,2:37,38:154,210:60:20:0.020,0.030,0.030:0.018,3.223e-03,0.979 0/0:75,0:0.020:45,0:30,0:36,0:154,0:0:0
GATK funcatator注释(要加interval文件)
在前面的mutect流程中,有提到了funcatator,现在我们就来实际使用,看看结果如何
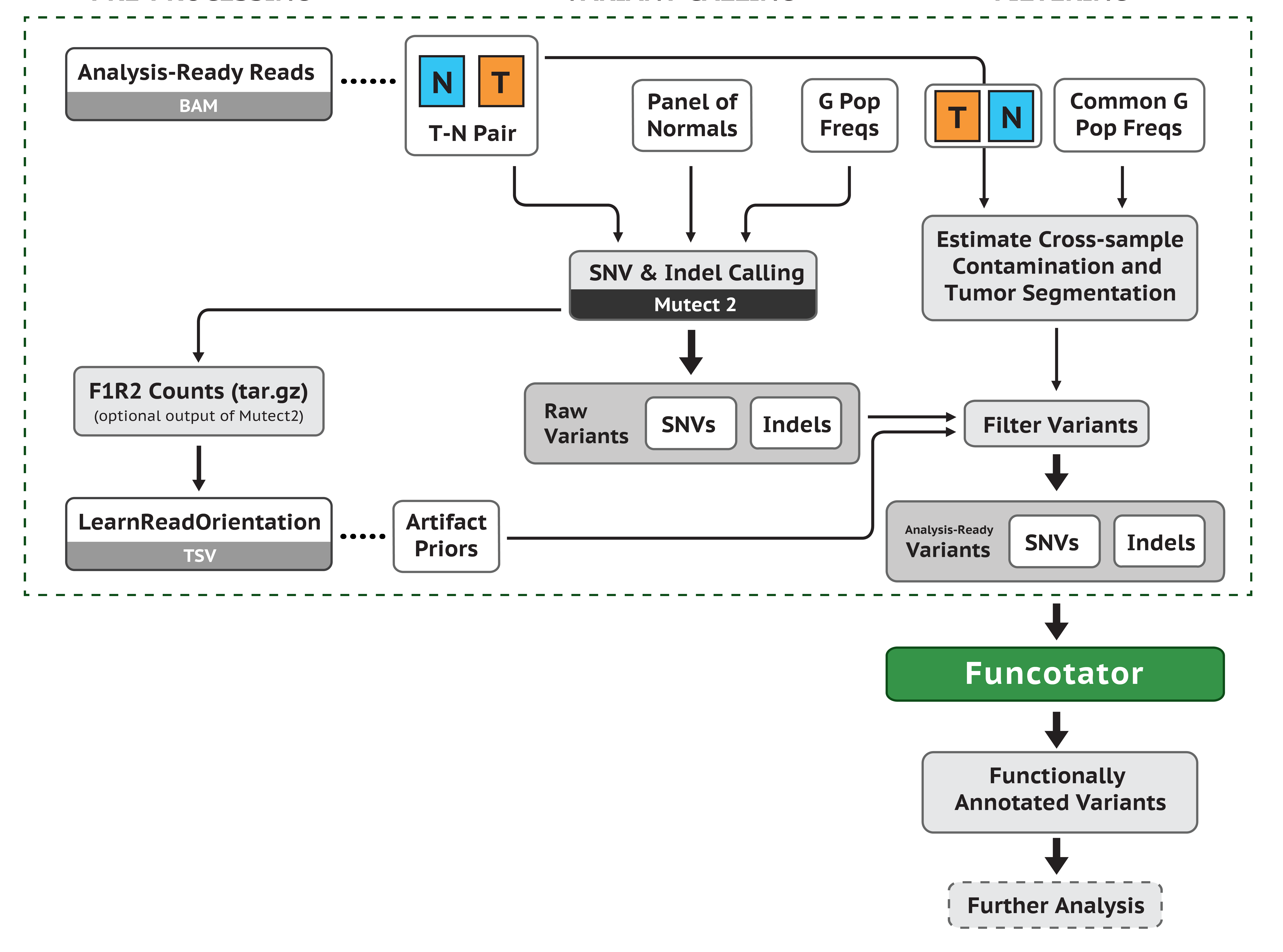
安装
注释只用到一个工具:Funcotator,帮助文档在:https://software.broadinstitute.org/gatk/documentation/tooldocs/4.1.4.1/org_broadinstitute_hellbender_tools_funcotator_Funcotator.php,需要注意的是,不同 GATK 版本所有到的参数可能不同,结果也不完全一致。这里我们使用最新版本的gatk-4.1.4.1的 Funcotator 工具来做注释,方法比较简单,首先需要下载数据库文件,前面本文已经下载过了,14G 左右的文件,解压后是 18G ,这里不再演示具体下载方法,简单来说就是下面代码:
cd ~/wes_cancer/datanohup wget -c ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/funcotator/funcotator_dataSources.v1.6.20190124s.tar.gz &tar -zxvf funcotator_dataSources.v1.6.20190124s.tar.gz
注释
然后使用GATK的Funcotator工具进行在线注释。
根据官网的教程https://software.broadinstitute.org/gatk/documentation/tooldocs/4.1.4.1/org_broadinstitute_hellbender_tools_funcotator_Funcotator.php,需要一个interval文件,interval文件和bed文件基本一致,只不过bed文件的坐标是从“0”开始编号,而interval_list文件是从“ 1”开始编号。这里我们已有bed文件,用gatk的BedToIntervalList工具可以转成interval:
GATK=~/wes_cancer/biosoft/gatk-4.1.4.1/gatkbed=~/wes_cancer/data/hg38.exon.beddict=~/wes_cancer/data/Homo_sapiens_assembly38.dict$GATK BedToIntervalList -I ${bed} -O hg38.exon.interval_list -SD ${dict}
准备好interval文件后回到工作目录
cd ~/wes_cancer/project
用到的脚本Funcotator.sh如下(经评论区读者指出,脚本需要添加对 vcf 创建索引,已更正):
GATK=~/wes_cancer/biosoft/gatk-4.1.4.1/gatkref=~/wes_cancer/data/Homo_sapiens_assembly38.fastainterval=~/wes_cancer/data//hg38.exon.interval_listsource=~/wes_cancer/data/funcotator_dataSources.v1.6.20190124scat config | while read iddoecho "start Funcotator for ${id} " `date`$GATK IndexFeatureFile --input ./6.mutect/${id}_filter.vcf$GATK Funcotator -R $ref \-V ./6.mutect/${id}_filter.vcf \-O ./7.annotation/funcotator/${id}_funcotator.tmp.maf \--data-sources-path ${source} \--intervals ${interval} \--output-file-format MAF \--ref-version hg38echo "end Funcotator for ${id} " `date`done
注释的速度也相当快,一个样本 10 秒左右就注释好了,同样我们以 case1_biorep_A_techrep 这个样本为例,打开查看一下结果,这里跳过了头文件:
$ less ./7.annotation/funcotator/case1_biorep_A_techrep_funcotator.maf | grep -v '^#' | head -2Hugo_Symbol Entrez_Gene_Id Center NCBI_Build Chromosome Start_Position End_Position Strand Variant_Classification Variant_Type Reference_Allele Tumor_Seq_Allele1 Tumor_Seq_Allele2 dbSNP_RS dbSNP_Val_Status Tumor_Sample_Barcode Matched_Norm_Sample_Barcode Match_Norm_Seq_Allele1 Match_Norm_Seq_Allele2 Tumor_Validation_Allele1 Tumor_Validation_Allele2 Match_Norm_Validation_Allele1 Match_Norm_Validation_Allele2 Verification_Status Validation_Status Mutation_Status Sequencing_Phase Sequence_Source Validation_Method Score BAM_File Sequencer Tumor_Sample_UUID Matched_Norm_Sample_UUID Genome_Change Annotation_Transcript Transcript_Strand Transcript_Exon Transcript_Position cDNA_Change Codon_ChangProtein_Change Other_Transcripts Refseq_mRNA_Id Refseq_prot_Id SwissProt_acc_Id SwissProt_entry_Id Description UniProt_AApos UniProt_Region UniProt_Site UniProt_Natural_Variations UniProt_Experimental_Info GO_Biological_Process GO_Cellular_Component GO_Molecular_Function COSMIC_overlapping_mutations COSMIC_fusion_genes COSMIC_tissue_types_affected COSMIC_total_alterations_in_gene Tumorscape_Amplification_Peaks Tumorscape_Deletion_Peaks TCGAscape_Amplification_Peaks TCGAscape_Deletion_Peaks DrugBank ref_context gc_content CCLE_ONCOMAP_overlapping_mutations CCLE_ONCOMAP_total_mutations_in_gene CGC_Mutation_Type CGC_Translocation_Partner CGC_Tumor_Types_Somatic CGC_Tumor_Types_Germline CGC_Other_Diseases DNARepairGenes_Activity_linked_to_OMIM FamilialCancerDatabase_Syndromes MUTSIG_Published_Results OREGANNO_ID OREGANNO_Values tumor_f t_alt_count t_ref_count n_alt_count n_ref_count Gencode_27_secondaryVariantClassification ACMGLMMLof_LOF_Mechanism ACMGLMMLof_Mode_of_Inheritance ACMGLMMLof_Notes ACMG_recommendation_Disease_Name ClinVar_VCF_AF_ESP ClinVar_VCF_AF_EXAC ClinVar_VCF_AF_TGP ClinVar_VCF_ALLELEID ClinVar_VCF_CLNDISDB ClinVar_VCF_CLNDISDBINCL ClinVar_VCF_CLNDN ClinVar_VCF_CLNDNINCL ClinVar_VCF_CLNHGVS ClinVar_VCF_CLNREVSTAT ClinVar_VCF_CLNSIG ClinVar_VCF_CLNSIGCONF ClinVar_VCF_CLNSIGINCL ClinVar_VCF_CLNVC ClinVar_VCF_CLNVCSO ClinVar_VCF_CLNVI ClinVar_VCF_DBVARID ClinVar_VCF_GENEINFO ClinVar_VCF_MC ClinVar_VCF_ORIGIN ClinVar_VCF_RS ClinVar_VCF_SSR ClinVar_VCF_ID ClinVar_VCF_FILTER LMMKnown_LMM_FLAGGED LMMKnown_ID LMMKnown_FILTER DP ECNT IN_PON NLOD N_ART_LOD POP_AF P_CONTAM P_GERMLINE RPA RU STR TLODCHD5 __UNKNOWN__ __UNKNOWN__ hg38 chr1 6146376 6146376 + Nonsense_Mutation SNP G G T __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN____UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ NA NA __UNKNOWN__ __UNKNOWN____UNKNOWN__ g.chr1:6146376G>T ENST00000262450.7 - 11 173c.1638C>A c.(1636-1638)tgC>tgA p.C546* __UNKNOWN__ __UNKNOWN____UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN____UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ CATCCATGTCGTTCTTTCTTT 0.5860349127182045 __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN____UNKNOWN__ __UNKNOWN__ 0.058 3 97 0 75 false 184 1 22.56 -1.888e+00 1.000e-05 0.00 -4.514e+01 5.41
可以看到这个工具一下子注释了几十个数据库,有一百多列的信息,当然很多注释结果是 __UNKNOWN__。
vep注释
在以前如要要用 vep 的话,安装比较麻烦,甚至需要管理员权限:http://www.bio-info-trainee.com/1600.html
也可以参考ensemble数据库对应的安装教程:http://asia.ensembl.org/info/docs/tools/vep/script/vep_download.html
强烈推荐,使用 conda 来安装 vep ,因为下面的教程写的比较早,就不修改了。
里面介绍了多种方法,最后一种是用 docker 装 vep,比较麻烦一点,但是也不难,需要学习一下 docker,也是需要管理员权限,这个方法的好处在于,docker 的 vep 镜像中已经把各种模块配置好了,启动镜像就可以用 vep注释,不需要下载,如果你有管理员权限,可以尝试这种方法。如果没有管理员权限,也可以购买一台配置较低的阿里云服务器(如果是学生还有优惠套餐,9.5 元/月)。对于这台服务器,你就有管理员权限啦,安装方法见上面的链接,这里就不再演示了(其实我也有试了其他几种方法的,但是最后都因为 perl 安装依赖模块缺失而放弃)
安装docker
sudo apt install docker-cedocker --versionsudo add-apt-repository "deb [arch=amd64] https://download.docker.com/linux/ubuntu artful stable"sudo docker run hello-world
用docker安装vep镜像
docker pull ensemblorg/ensembl-vepdocker run -t -i ensemblorg/ensembl-vep ./vep# Create a directory on your machine:mkdir $HOME/vep_data# Make sure that the created directory on your machine has read and write access granted# so the docker container can write in the directory (VEP output):chmod a+rwx $HOME/vep_datadocker run -t -i -v $HOME/vep_data:/opt/vep/.vep ensemblorg/ensembl-vepdocker run -t -i -v $HOME/vep_data:/opt/vep/.vep ensemblorg/ensembl-vep perl INSTALL.pl -a cfp -s homo_sapiens -y GRCh38 -g all
启动镜像
根据上面的教程,我们在家目录中新建了一个vep_data的文件夹,然后把我们需要注释的vcf文件拷贝到vep_data目录下,然后启动vep镜像:
sudo docker run -t -i -v $HOME/vep_data:/opt/vep/.vep ensemblorg/ensembl-vep
注释
用vep注释的脚本vep.sh如下:
cat /opt/vep/.vep/wes_cancer/project/config | while read iddoecho "start vep_annotation for ${id} " `date`./vep --cache --offline --format vcf --vcf --force_overwrite \--dir_cache /opt/vep/.vep/ \--dir_plugins /opt/vep/.vep/Plugins/ \--input_file /opt/vep/.vep/wes_cancer/project/6.mutect/${id}_filter.vcf \--output_file /opt/vep/.vep/wes_cancer/project/7.annotation/vep/${id}_vep.vcfecho "end vep_annotation for ${id} " `date`done
注释后结果,每个样本会输出两个文件,一个是vcf文件,一个是html文件,如:
case1_biorep_A_techrep_vep.vcfcase1_biorep_A_techrep_vep.vcf_summary.html
同样的,我们查看一下样本case1_biorep_A_techrep_vep.vcf注释的结果,可以发现,vep的注释结果就是在原先的vcf文件的INFO列后面注释上了很多的信息,这些信息以|符号作为分隔
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT case1_biorep_A_techrep case1_germlinechr1 6146376 . G T . PASS DP=184;ECNT=1;NLOD=22.56;N_ART_LOD=-1.888e+00;POP_AF=1.000e-05;P_CONTAM=0.00;P_GERMLINE=-4.514e+01;TLOD=5.41;CSQ=T|missense_variant|MODERATE|CHD5|ENSG00000116254|Transcript|ENST00000262450|protein_coding|11/42||||1936|1638|546|N/K|aaC/aaA|||-1||HGNC|HGNC:16816,T|upstream_gene_variant|MODIFIER|CHD5|ENSG00000116254|Transcript|ENST00000462991|nonsense_mediated_decay|||||||||||2272|-1|cds_start_NF|HGNC|HGNC:16816,T|missense_variant&NMD_transcript_variant|MODERATE|CHD5|ENSG00000116254|Transcript|ENST00000496404|nonsense_mediated_decay|11/34||||1638|1638|546|N/K|aaC/aaA|||-1||HGNC|HGNC:16816 GT:AD:AF:F1R2:F2R1:MBQ:MFRL:MMQ:MPOS:SA_MAP_AF:SA_POST_PROB 0/1:97,3:0.058:48,1:49,2:37,38:154,210:60:20:0.020,0.030,0.030:0.018,3.223e-03,0.979 0/0:75,0:0.020:45,0:30,0:36,0:154,0:0:0
SNPEFF注释
安装
snpeff的安装方法也比较简单,参考:
http://snpeff.sourceforge.net/download.html#source
cd ~/wes_cancer/biosoft# Download latest versionwget http://sourceforge.net/projects/snpeff/files/snpEff_latest_core.zip# Unzip fileunzip snpEff_latest_core.zip
对于数据库文件,我们需要下载人类参考基因组 hg38 版本的注释文件 Homo_sapiens,但是:By default SnpEff automatically downloads and installs the database for you, so you don’t need to do it manually. 软件默认下载了 GRCh38.86 ,所以我们不需要自己下载,如果需要其他数据库,可以通过下面命令查询和获取下载的链接,有 20000 多个 database,足够了,不过需要注意一下版本的问题:
java -jar snpEff.jar databases | less -S
注释
对于癌症样本,有其特定的注释方法:
http://snpeff.sourceforge.net/SnpEff_manual.version_4_0.html#cancer
需要一个 samples_cancer.txt 文件,内容如下:
$ cat ~/wes_cancer/biosoft/snpeff/samples_cancer.txtcase1_germline case1_biorep_A_techrepcase2_germline case2_biorep_Acase3_germline case3_biorep_Acase4_germline case4_biorep_Acase5_germline case5_biorep_A......
注释的脚本snpeff.sh如下
dir=~/wes_cancer/project/7.annotation/config=~/wes_cancer/project/configcat ${config} | while read iddoecho "start snpeff_annotation for ${id} " `date`java -Xmx4g -jar snpEff.jar -v \-cancer -cancerSamples ./7.annotation/snpeff/samples_cancer.txt \GRCh38.86 \./6.mutect/${id}_filter.vcf > ./7.annotation/snpeff/${id}_eff.vcfecho "end snpeff_annotation for ${id} " `date`done
这个运行速度比较慢一些,一个样本大概需要一分钟的时间,我们还是来看一下注释的结果,同样以这个样本case1_biorep_A_techrep为例:
$ head -1 case1_biorep_A_techrep_eff.vcf
chr1 6461445 . G T . PASS DP=21;ECNT=1;NLOD=2.70;N_ART_LOD=-1.049e+00;POP_AF=1.000e-05;P_CONTAM=0.00;P_GERMLINE=-1.853e+00;TLOD=6.33;ANN=T|missense_variant|MODERATE|TNFRSF25|ENSG00000215788|transcript|ENST00000377782.7|protein_coding|10/10|c.1270C>A|p.Arg424Ser|1338/1632|1270/1281|424/426||,T|missense_variant|MODERATE|TNFRSF25|ENSG00000215788|transcript|ENST00000356876.7|protein_coding|10/10|c.1243C>A|p.Arg415Ser|1331/1625|1243/1254|415/417||,T|missense_variant|MODERATE|TNFRSF25|ENSG00000215788|transcript|ENST00000351959.9|protein_coding|9/9|c.1132C>A|p.Arg378Ser|1200/1441|1132/1143|378/380||,T|missense_variant|MODERATE|TNFRSF25|ENSG00000215788|transcript|ENST00000348333.7|protein_coding|9/9|c.1108C>A|p.Arg370Ser|1108/1119|1108/1119|370/372||,T|missense_variant|MODERATE|TNFRSF25|ENSG00000215788|transcript|ENST00000351748.7|protein_coding|5/5|c.694C>A|p.Arg232Ser|694/705|694/705|232/234||,T|3_prime_UTR_variant|MODIFIER|TNFRSF25|ENSG00000215788|transcript|ENST00000480393.5|nonsense_mediated_decay|9/9|c.*582C>A|||||1463|,T|3_prime_UTR_variant|MODIFIER|TNFRSF25|ENSG00000215788|transcript|ENST00000485036.5|nonsense_mediated_decay|9/9|c.*530C>A|||||1215|,T|3_prime_UTR_variant|MODIFIER|TNFRSF25|ENSG00000215788|transcript|ENST00000414040.6|nonsense_mediated_decay|9/9|c.*530C>A|||||1215|,T|3_prime_UTR_variant|MODIFIER|TNFRSF25|ENSG00000215788|transcript|ENST00000502588.5|nonsense_mediated_decay|7/7|c.*530C>A|||||1215|,T|3_prime_UTR_variant|MODIFIER|TNFRSF25|ENSG00000215788|transcript|ENST00000502730.5|nonsense_mediated_decay|5/5|c.*446C>A|||||677|,T|3_prime_UTR_variant|MODIFIER|TNFRSF25|ENSG00000215788|transcript|ENST00000510563.5|nonsense_mediated_decay|8/8|c.*530C>A|||||1215|,T|downstream_gene_variant|MODIFIER|ESPN|ENSG00000187017|transcript|ENST00000377828.5|protein_coding||c.*1299G>T|||||501|,T|downstream_gene_variant|MODIFIER|ESPN|ENSG00000187017|transcript|ENST00000461727.5|protein_coding||c.*1299G>T|||||516|,T|downstream_gene_variant|MODIFIER|TNFRSF25|ENSG00000215788|transcript|ENST00000461703.6|processed_transcript||n.*2091C>A|||||2091|,T|downstream_gene_variant|MODIFIER|TNFRSF25|ENSG00000215788|transcript|ENST00000453341.1|retained_intron||n.*2666C>A|||||2666|,T|downstream_gene_variant|MODIFIER|PLEKHG5|ENSG00000171680|transcript|ENST00000537245.5|protein_coding||c.*6118C>A|||||4647|,T|downstream_gene_variant|MODIFIER|ESPN|ENSG00000187017|transcript|ENST00000477679.1|retained_intron||n.*1034G>T|||||1034|,T|downstream_gene_variant|MODIFIER|ESPN|ENSG00000187017|transcript|ENST00000468561.1|processed_transcript||n.*75G>T|||||75|,T|downstream_gene_variant|MODIFIER|TNFRSF25|ENSG00000215788|transcript|ENST00000453260.6|retained_intron||n.*35C>A|||||35|,T|downstream_gene_variant|MODIFIER|PLEKHG5|ENSG00000171680|transcript|ENST00000489097.5|retained_intron||n.*4647C>A|||||4647|,T|downstream_gene_variant|MODIFIER|PLEKHG5|ENSG00000171680|transcript|ENST00000535355.5|protein_coding||c.*6118C>A|||||4647|,T|downstream_gene_variant|MODIFIER|PLEKHG5|ENSG00000171680|transcript|ENST00000377748.5|protein_coding||c.*6118C>A|||||4647|,T|downstream_gene_variant|MODIFIER|ESPN|ENSG00000187017|transcript|ENST00000434576.1|protein_coding||c.*260G>T|||||75|WARNING_TRANSCRIPT_NO_START_CODON,T|downstream_gene_variant|MODIFIER|TNFRSF25|ENSG00000215788|transcript|ENST00000475730.5|processed_transcript||n.*311C>A|||||311|,T|downstream_gene_variant|MODIFIER|TNFRSF25|ENSG00000215788|transcript|ENST00000469691.5|retained_intron||n.*549C>A|||||549|,T|downstream_gene_variant|MODIFIER|TNFRSF25|ENSG00000215788|transcript|ENST00000515145.1|retained_intron||n.*3122C>A|||||3122|,T|downstream_gene_variant|MODIFIER|ESPN|ENSG00000187017|transcript|ENST00000416731.5|protein_coding||c.*1299G>T|||||1047|,T|downstream_gene_variant|MODIFIER|ESPN|ENSG00000187017|transcript|ENST00000475228.5|protein_coding||c.*1393G>T|||||1393|WARNING_TRANSCRIPT_INCOMPLETE,T|downstream_gene_variant|MODIFIER|ESPN|ENSG00000187017|transcript|ENST00000633239.1|protein_coding||c.*1299G>T|||||519|WARNING_TRANSCRIPT_NO_START_CODON,T|downstream_gene_variant|MODIFIER|TNFRSF25|ENSG00000215788|transcript|ENST00000481401.5|protein_coding||c.*1192C>A|||||1192|WARNING_TRANSCRIPT_INCOMPLETE,T|downstream_gene_variant|MODIFIER|PLEKHG5|ENSG00000171680|transcript|ENST00000400913.5|protein_coding||c.*6118C>A|||||4647|,T|downstream_gene_variant|MODIFIER|PLEKHG5|ENSG00000171680|transcript|ENST00000340850.9|protein_coding||c.*6118C>A|||||4647|,T|downstream_gene_variant|MODIFIER|ESPN|ENSG00000187017|transcript|ENST00000636330.1|protein_coding||c.*5248G>T|||||4774|,T|downstream_gene_variant|MODIFIER|ESPN|ENSG00000187017|transcript|ENST00000636644.1|protein_coding||c.*4182G>T|||||4182|WARNING_TRANSCRIPT_INCOMPLETE,T|non_coding_transcript_exon_variant|MODIFIER|TNFRSF25|ENSG00000215788|transcript|ENST00000473343.5|retained_intron|4/4|n.1297C>A||||||,T|non_coding_transcript_exon_variant|MODIFIER|TNFRSF25|ENSG00000215788|transcript|ENST00000513135.5|retained_intron|6/6|n.3189C>A|||||| GT:AD:AF:F1R2:F2R1:MBQ:MFRL:MMQ:MPOS:SA_MAP_AF:SA_POST_PROB 0/1:6,2:0.278:3,2:3,0:29,39:204,184:60:14:0.00,0.253,0.250:0.023,0.025,0.952 0/0:9,0:0.126:3,0:6,0:24,0:169,0:0:0
同样是在原来的 vcf 文件的INFO列添加了许多注释信息,这里仅仅是该样本的第一个 vcf 突变位点,就注释上了大量的信息,主要是这个位点刚好处于多个基因上,且每个基因有多个转录本。这个结果和 vep 注释后拿到的结果非常相似。

