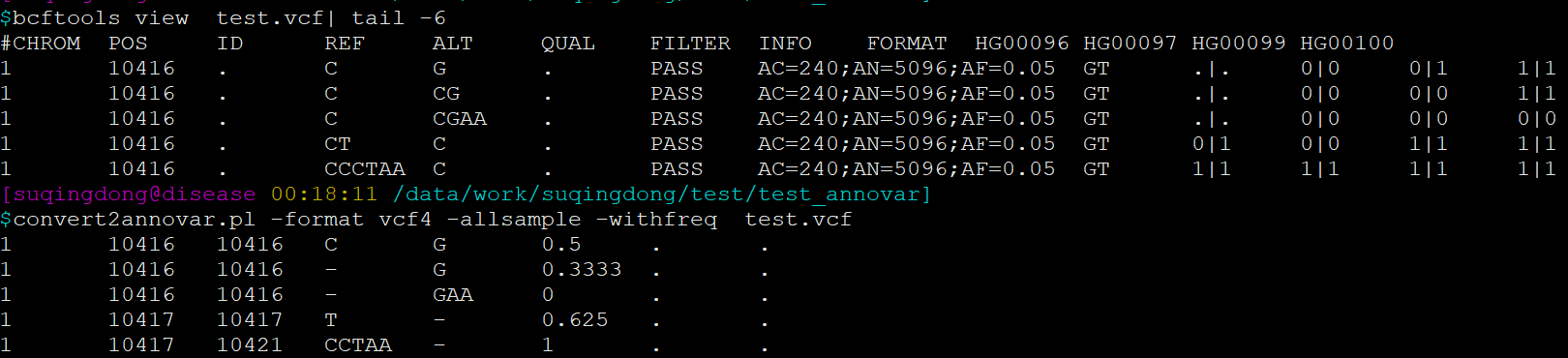
# for SNPSTART = END = POSREF = REFALT = ALT# for INSSTART = END = POSREF = '-'ALT = ALT[1:]# for DELSTART = POS + 1END = POS + len(REF) - 1REF = REF[1:]ALT = '-'
PS:多个样本的VCF转换时需要添加参数
--allsample --withfreq频率会根据GT重新计算AF = AC / ANAC: 突变的Allele数,0/1记为1, 1/1记为2 AN:发生突变的Allele数,./.记为0,0/0,0/1,1/1均记为2(即除了./.的样本数的2倍)

