由於語法渲染問題而影響閱讀體驗, 請移步博客閱讀~
本文GitPage地址
RNA-seq with Trinity
Downloading RNA-Seq sequences
For this practicing, I found a group of mouse RNA-Seq which have replicates and the size of SRA is easy handle for your PC.
SRR14962733 to SRR149627346
prefetch SRR14962733prefetch SRR14962734prefetch SRR14962735prefetch SRR14962736# orfor i in {3..6};do prefetch SRR1496273$i;done
download with ascp
##### Downloading the RNA-seqprefetch --ascp-path "/usr/bin/ascp|/home/ken/.aspera/connect/etc/asperaweb_id_dsa.putty" ERR025599
Seq Clean
Turn sra to fastq
## move the result from default directory to local
mv ~/ncbi/public/sra .
##### Split SSR to fastq
fastq-dump --defline-seq '@$sn[_$rn]/$ri' --split-files sra/*
mkdir fastq
mv *fastq fastq
Quality control
SRA download from NCBI doesn’t included the ASCII grades of the reads now. So, we can’t doing this for free-download reads. I’ll skip this step.
##### cut the low quality reads
##basic command
##fastp -u 15 -w 8 -i SRR7760055_1.fastq -o cut_SRR7760055_1.fastq
for i in $(ls *.fastq);do fastp -u 15 -w 8 -i $i -o cut_$i; mv fastp.html $i.html;done
##### Trimmomatic cut the lower grade head or tail jodged by the quality report
java -jar trimmomatic-0.38.jar SE -threads 8 -phred33 SRR771602.fastq 2.fq ILLUMINACLIP:TruSeq3-SE:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36 HEADCROP:10 CROP:87
for i in $(ls cut_SRR77600*);do
java -jar trimmomatic-0.38.jar SE -threads 8 -phred33 $i 2_$i ILLUMINACLIP:TruSeq3-SE:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36 HEADCROP:10 CROP:87
done
De-novo
We recommend generating a single Trinity assembly based on combining all reads across all samples as inputs.
cite: Tirnity wiki
Now, waiting…
# merge all reads
cat fastq/* > all.fastq
##Trnity
Time1=$(date)
Trinity --seqType fq --max_memory 55G --single all.fastq --CPU 8 --full_cleanup
Time2=$(date)
It takes me 81 min.
errors
Error, do not understand options: fastq/SRR14962734_1.fastq
Result
I made a mistake. With the codes above, I got only three sequences. So, I suspect that mabey this is not RNA-Seq but miRNA-seq. I looked back to the NCBI and checking carefully this time. Guess what? This is a miRNA-Seq! = =
You can find the information in:
Library:
Name: …
Strategy: miRNA-Seq
Now, let’s repeat the codes above with another groups: SRR14962739-SRR14962742
# or
for i in {39..42};
do prefetch SRR149627$i;
done
check the time consuming:
du all.fastq
echo $Time1
echo $Time2
14G all.fastq
Sun Jul 4 06:20:58 PM CST 2021
Mon Jul 5 02:36:32 AM CST 2021
about 8 hours
Statistical information of the result
Trinity supplied a perl script to calculating and evaluating the result: TrinityStats.pl
It has two group of results:
- Stats based on ALL transcript contigs:
- Stats based on ONLY LONGEST ISOFORM per ‘GENE’:
{$path}/trinityrnaseq-v2.12.0/util/TrinityStats.pl trinity_out_dir.Trinity.fasta
################################
## Counts of transcripts, etc.
################################
Total trinity 'genes': 41400
Total trinity transcripts: 42451
Percent GC: 49.50
########################################
Stats based on ALL transcript contigs:
########################################
Contig N10: 6068
Contig N20: 4371
Contig N30: 3377
Contig N40: 2687
Contig N50: 2100
Median contig length: 426
Average contig: 991.80
Total assembled bases: 42102797
#####################################################
## Stats based on ONLY LONGEST ISOFORM per 'GENE':
#####################################################
Contig N10: 5781
Contig N20: 4155
Contig N30: 3202
Contig N40: 2543
Contig N50: 1982
Median contig length: 415
Average contig: 948.27
Total assembled bases: 39258319
Downstream Analysis
We are processing the downstream analysis based on the scripts trinity supplied. Most of them are in the ${Trinity directory}/util/
TrinityStats.pl
abundance_estimates_to_matrix.pl
align_and_estimate_abundance.pl
analyze_blastPlus_topHit_coverage.pl
filter_low_expr_transcripts.pl
insilico_read_normalization.pl
retrieve_sequences_from_fasta.pl
sift_bam_max_cov.pl
TrinityStats.pl is for calculating the value of N50
Align and Abundance Estimate
The script, align_and_estimate_abundance.pl, was used here to estimate the abundances of each transcripts from your transcriptome library. More details about this scripts is in trinity wiki
Because we have replicates, we need a file to interpreted the groups. (group.csv)
miR-31 miR-31_rep1 fastq/SRR14962739_1.fastq
miR-31 miR-31_rep2 fastq/SRR14962740_1.fastq
Scr-2 Scr-2_rep1 fastq/SRR14962741_1.fastq
Scr-2 Scr-2_rep2 fastq/SRR14962742_1.fastq
```bash
~/Bio/trinityrnaseq-v2.12.0/util/align_and_estimate_abundance.pl --transcripts trinity_out_dir.Trinity.fasta --seqType fq --samples_file group.csv --est_method RSEM --aln_method bowtie --trinity_mode --prep_reference --output_dir rsem_outdir
It could build index, pair the reads and counting the matched reads automatically.
After down, you’ll got a bunch of result. The abaundance file was in the directories miR-31_rep1, miR-31_rep2, Scr-2_rep1, Scr-2_rep2 as the statistic scripts, the result was divided into to groups, gene and isoforms: RSEM.genes.results RSEM.isoforms.results. This is where the count, TPM, and FPKM were generated and stored (If you are using RSEM).
FPKM: fragments per kilobase transcript length per million fragments mapped
TPM: transcripts per million transcripts
# Check the results.
head miR-31_rep1/RSEM.isoforms.results
transcript_id gene_id length effective_length expected_count TPM FPKM IsoPct
TRINITY_DN10000_c0_g1_i1 TRINITY_DN10000_c0_g1 3506 3457.00 489.00 10.97 6.58 100.00
TRINITY_DN10001_c0_g1_i1 TRINITY_DN10001_c0_g1 441 392.00 15.00 2.97 1.78 100.00
TRINITY_DN10002_c0_g1_i1 TRINITY_DN10002_c0_g1 5392 5343.00 2609.00 37.85 22.73 100.00
TRINITY_DN10003_c0_g1_i1 TRINITY_DN10003_c0_g1 1419 1370.00 1995.00 112.89 67.78 100.00
TRINITY_DN10004_c0_g1_i1 TRINITY_DN10004_c0_g1 3829 3780.00 1715.00 35.17 21.12 100.00
TRINITY_DN10005_c0_g1_i1 TRINITY_DN10005_c0_g1 2379 2330.00 1755.00 58.39 35.06 100.00
Expression matrix
Of course that trinity supplied a script, abundance_estimates_to_matrix.pl, do it for you as well.
~/Bio/trinityrnaseq-v2.12.0/util/abundance_estimates_to_matrix.pl --est_method RSEM \
--gene_trans_map trinity_out_dir.Trinity.fasta.gene_trans_map \
--out_prefix RSEM \
--name_sample_by_basedir \
Scr-2_rep1/RSEM.isoforms.results \
Scr-2_rep2/RSEM.isoforms.results \
miR-31_rep1/RSEM.isoforms.results \
miR-31_rep2/RSEM.isoforms.results
# my results of prefix is RSEM
ls RSEM*
RSEM.gene.TMM.EXPR.matrix RSEM.gene.counts.matrix RSEM.isoform.TPM.not_cross_norm.runTMM.R
RSEM.gene.TPM.not_cross_norm RSEM.isoform.TMM.EXPR.matrix RSEM.isoform.counts.matrix
RSEM.gene.TPM.not_cross_norm.TMM_info.txt RSEM.isoform.TPM.not_cross_norm
RSEM.gene.TPM.not_cross_norm.runTMM.R RSEM.isoform.TPM.not_cross_norm.TMM_info.txt
Now, all the matrix were extracted from separated results of samples.
move them into a new directory
mkdir RSEM_result
mv RSEM* RSEM_result
Errors
- libreadline
/home/ken/miniconda3/envs/Biostation/lib/R/bin/exec/R: error while loading shared libraries: libreadline.so.6: cannot open shared object file: No such file or directory
chech the libreadlines:
ls /lib/libreadline.*
/lib/libreadline.so /lib/libreadline.so.8 /lib/libreadline.so.8.1
The problem is this script is relying on an very old version. So, you can heck on the script, or cheat it.
This is how I fool the script:
sudo ln -s /lib/libreadline.so.8 /lib/libreadline.so.6
- libncurses
/home/ken/miniconda3/envs/Biostation/lib/R/bin/exec/R: error while loading shared libraries: libncurses.so.5: cannot open shared object file: No such file or directory
The same as error 1
sudo ln -s /lib/libncursesw.so.6 /lib/libncurses.so.5
Low-Expression Reads Filter
The results of the matrix are huge. we got 42451 pieces of isforms but lot’s of them are low expression gene we want to remove. You can achive this by using R. Trinity supplied the related scripts to handle with this, too.
This is the function in R to down the same thing as the scripts from Trinity
A <- read.table("RSEM.isoform.TPM.not_cross_norm")
Filter <- function(A, Th){
tmp = A
for(i in c(1:ncol(A))){
tmp = tmp[which(tmp[,i] < Th),]
}
return(tmp)
}
In trinity, perl scripts, util/misc/count_matrix_features_given_MIN_TPM_threshold.pl, is aiming to that the genes/isoforms expressed above a minimum TPM expression threshold in any sample.
~/Bio/trinityrnaseq-v2.12.0/util/misc/count_matrix_features_given_MIN_TPM_threshold.pl \
RSEM.isoform.TPM.not_cross_norm
neg_min_tpm num_features
-6771 1
-6758 2
-6185 3
-6092 4
.
.
.
-4 36909
-3 41004
-2 42353
-1 42440
0 42451
In this result, -1 42440 means that 42440 isoforms’s TPM is larger than 1 in all samples. You can get the same result with the function in R above. (Filter(A, 1))
Ploting the result
# write the result into a file
~/Bio/trinityrnaseq-v2.12.0/util/misc/count_matrix_features_given_MIN_TPM_threshold.pl RSEM.isoform.TPM.not_cross_norm| tee isoform_matrix.TPM.not_cross_norm.counts_by_min_TPM
Plot the result with R
data = read.table("isoform_matrix.TPM.not_cross_norm.counts_by_min_TPM", header=T)
plot(data, xlim=c(-100,0), ylim=c(0,100000), t='b')

Filtering
By the function above, you can easily filtering the matrix from the row matrix. Trinity also supplied a pl script to extracting the transcripts from your transcriptome (you fasta file) to build a new library.
~/Bio/trinityrnaseq-v2.12.0/util/filter_low_expr_transcripts.pl \
--matrix RSEM.isoform.TPM.not_cross_norm \
--transcripts ../trinity_out_dir.Trinity.fasta \
--min_expr_any 5
Expression matrix QC
Checking the replicates
relied R packages: fastcluster,
~/Bio/trinityrnaseq-v2.12.0/Analysis/DifferentialExpression/PtR \
--matrix RSEM_result/RSEM.isoform.counts.matrix \
--samples group.csv --log2 --CPM \
--min_rowSums 10 \
--compare_replicates
Processing replicate QC analysis for sample: miR-31
for plotting:: min.raw: 0.994299466204194 max.raw: 1
Processing replicate QC analysis for sample: Scr-2
for plotting:: min.raw: 0.993762297454055 max.raw: 1
You’ll get the PDF result like it’s show in Trnity wiki
Checking all groups
~/Bio/trinityrnaseq-v2.12.0/Analysis/DifferentialExpression/PtR \
--matrix RSEM_result/RSEM.isoform.counts.matrix \
--samples group.csv --log2 --CPM \
--min_rowSums 10 \
--sample_cor_matrix
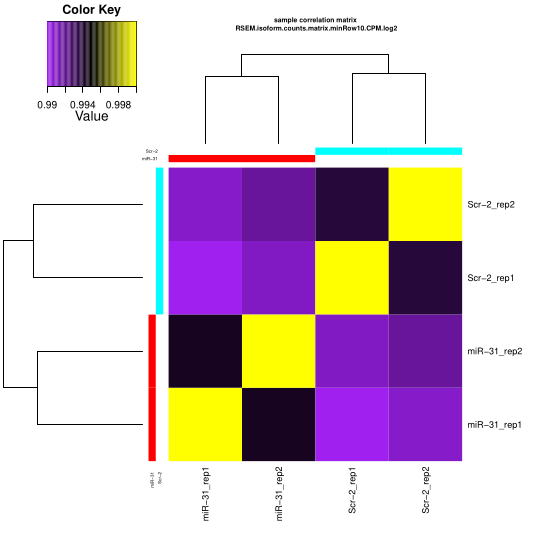
PCA Analysis
~/Bio/trinityrnaseq-v2.12.0/Analysis/DifferentialExpression/PtR \
--matrix RSEM_result/RSEM.isoform.counts.matrix \
--samples group.csv --min_rowSums 10 --log2 \
--CPM --center_rows \
--prin_comp 3
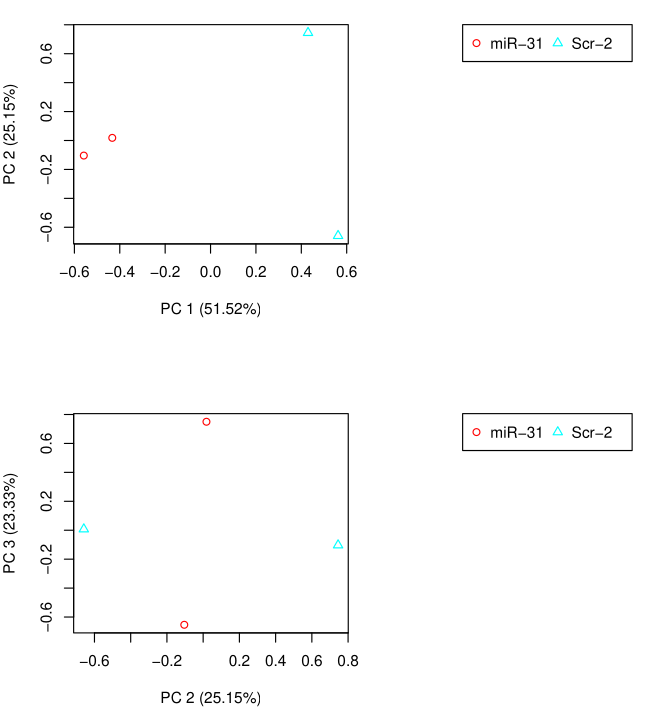
Differential Expression Matrix
Here we are supposed to using counts file for calculating.
~/Bio/trinityrnaseq-v2.12.0/Analysis/DifferentialExpression/run_DE_analysis.pl \
--matrix RSEM_result/RSEM.gene.counts.matrix \
--method voom \
--samples_file group.csv
a voom.******.dir directory was generated by default.
You can also using edgeR/DESeq2 instead of voom
Extracting and clustering differentially expressed transcripts
Now, enter the directory generated above and run codes below
~/Bio/trinityrnaseq-v2.12.0/Analysis/DifferentialExpression/analyze_diff_expr.pl\
--matrix ../RSEM_result/RSEM.gene.TMM.EXPR.matrix -P 1e-3 -C 2
Now, you can have the ugly heatmap ^_^
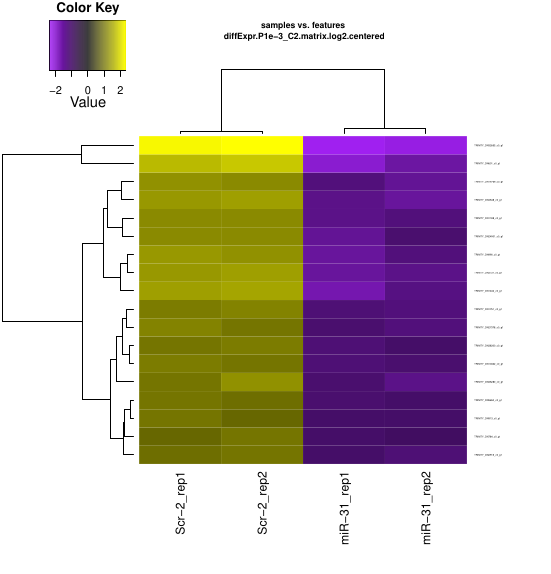
I tried those three different algorithms and get different results.
| Packages | Number of DEGs |
|---|---|
| edgeR | 149 |
| DESeq2 | 167 |
| Voom | 18 |
Enjoy~
由於語法渲染問題而影響閱讀體驗, 請移步博客閱讀~
本文GitPage地址
GitHub: Karobben
Blog:Karobben
BiliBili:史上最不正經的生物狗

