 |
|---|
| © pymol |
由於語法渲染問題而影響閱讀體驗, 請移步博客閱讀~
本文GitPage地址
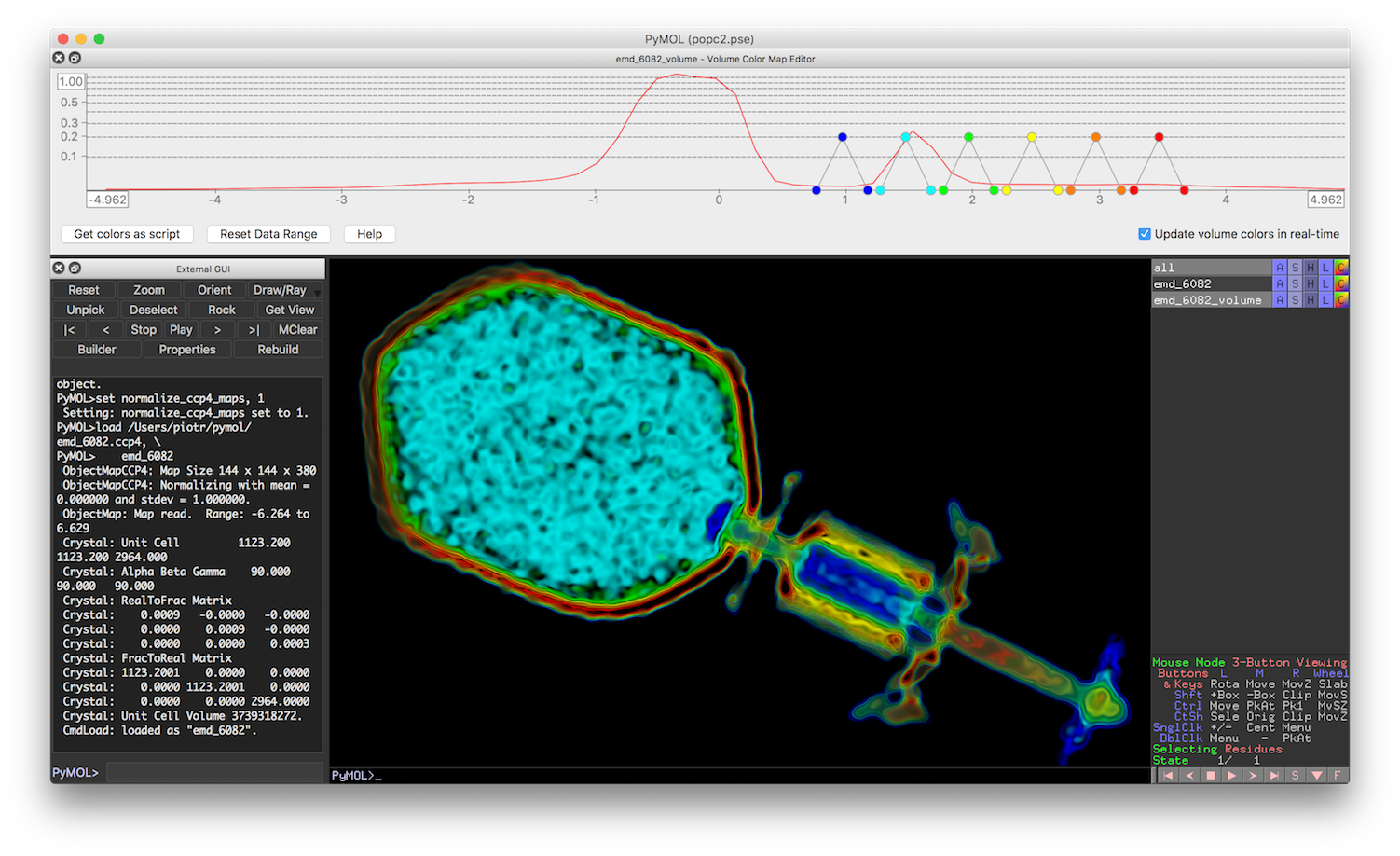
View and Themes
background color
bg_color grey30
Add something
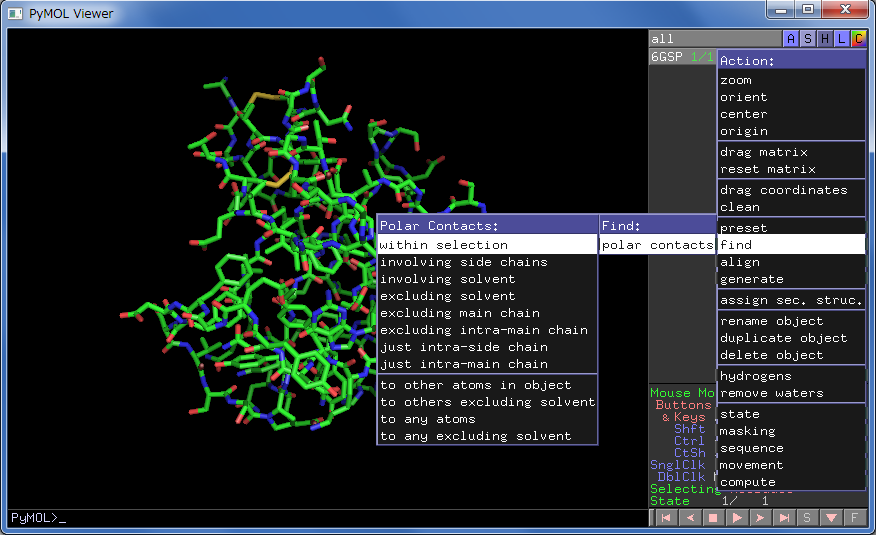
Add Hydrogen bonds
Add Hydrogen bonds: PyMOL tutorial
Action → find → polar contacts → select from menu
 |
|---|
| © PyMOL tutorial |
Remove something
cite: © Jan-Philip Gehrcke; 2011
# removing hydrogensremove (hydro)remove hydrogens# removing waterremove resn hoh# removing solventremove solvent
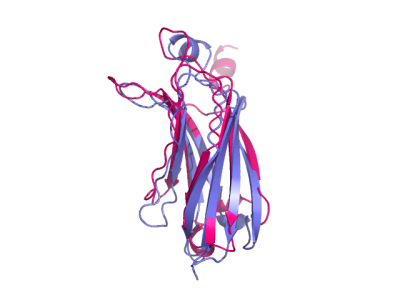
Strcture align
 |
|---|
| © Pymol |
fetch 1oky 1t46, async=0# 1) default with outlier rejectionalign 1oky, 1t46# 2) with alignment object, save to clustalw filealign 1oky, 1t46, object=alnobjsave alignment.aln, alnobj# 3) all-atom RMSD (no outlier rejection) and without superpositionalign 1oky, 1t46, cycles=0, transform=0
Partial structure align
Cite: Queen’s University
align 5cha and resi 1-100, 2xxl and resi 300-400
# or in short form:
align structure2 & i. 1-100, structure 1 & i. 300-400
# Furthermore, you may wish to restrict the alignment to just the backbone atoms, so you can say:
align structure2 and resi 1-100 and name n+ca+c+o, structure1 and resi 300-400 and name n+ca+c+o
# or in short form:
align structure2 & i. 1-100 & n. n+ca+c+o, structure1 & i. 300-400 & n. n+ca+c+o
Align chains
align 5cha and chain A+B+C, 2xxl and chain A
Atom
Atom color
## change the whole proteins color
color grey90, 2xxl
color grey80, 2xxl 5cha # 2 proteins
Select Atom
select aas, resn ASP+GLU in 2xxl
Create a variate ass which contain all ASP and GLU residues.
Enjoy~
由於語法渲染問題而影響閱讀體驗, 請移步博客閱讀~
本文GitPage地址
GitHub: Karobben
Blog:Karobben
BiliBili:史上最不正經的生物狗

