由於語法渲染問題而影響閱讀體驗, 請移步博客閱讀~
本文GitPage地址
Fasta Sequences Align
Quick look about the data
Suppose there is a fasta file name as LYSV-NCBI.fasta
Have a quick look
grep ">" LYSV-NCBI.fasta |wcwc LYSV-NCBI.fastaecho $(wc LYSV-NCBI.fasta| awk '{print $2}')-$(grep ">" LYSV-NCBI.fasta |wc |awk '{print $2}')|bc
124 1114 836318250 19116 1282572 LYSV-NCBI.fasta18002
As you can see, there are 124 sequences and roughly about 18002 bases
Let’s have a quick look at the sequences by Seq-view
Seq-view1.3 -i LYSV-NCBI.fasta
ClustelW2
Install
url: click me
wget http://www.clustal.org/download/current/clustalw-2.1.tar.gz
tar -zxvf clustalw-2.1.tar.gz
cd clustalw-2.1/
./configure
make
make install
clustalw2 -QUICKTREE -OUTPUT=FASTA -INFILE=LYSV-NCBI.fasta
It takes less than 20 mins

Quick look
Seq-view1.3 -i LYSV-NCBI.fasta -a 90

ALign prarmeters
UPGAM Tree
clustalw2 -QUICKTREE -OUTPUT=FASTA -INFILE=LYSV-NCBI.fasta -CLUSTERING=UPGMA -BOOTSTRAP=1000
clustalw2 -QUICKTREE -OUTPUT=FASTA -INFILE=LYSV-NCBI.fasta -CLUSTERING=NJ -BOOTSTRAP=1000
Muscle
apt install muscle
time muscle -in LYSV-NCBI.fasta -out 123.fa
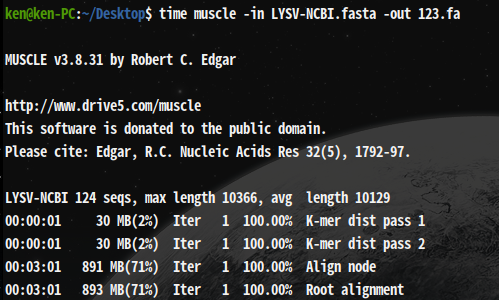
real 17m59.470s
user 17m59.034s
sys 0m0.256s
It takes about 18min as well as ClustelW2
Tcoffee
Source: T-Coffee
Protein sequences
# Default
t_coffee sample_seq1.fasta
# Quick
t_coffee sample_seq1.fasta -mode quickaln
# Consistent (M-Coffee combines the most common MSA packages)
t_coffee sample_seq1.fasta -mode mcoffee
# Structure (Expresso finds structures homologous to your sequences)
t_coffee sample_seq1.fasta -mode expresso
# Homology (PSI-Coffee enriches your dataset with homologous sequences)
t_coffee sample_seq1.fasta -mode psicoffee
# Accurate (combines Structures and Homology)
t_coffee sample_seq1.fasta -mode accurate
DNA sequences
# Default
t_coffee sample_dnaseq1.fasta
# Functional (Pro-Coffee increases accuracy of functional DNA regions )
t_coffee sample_dnaseq1.fasta -mode procoffee
RNA sequences
# Default
t_coffee sample_rnaseq1.fasta
# Structure 2D (R-Coffee uses predicted secondary structures)
t_coffee sample_rnaseq1.fasta -mode rcoffee
# Structure 3D (R-Coffee combined with Consan structural alignments)
t_coffee sample_rnaseq1.fasta -mode rcoffee_consan
# Accurate (RM-Coffee use M-Coffee and secondary structure predictions)
t_coffee sample_rnaseq1.fasta -mode rmcoffee
Enjoy~
由於語法渲染問題而影響閱讀體驗, 請移步博客閱讀~
本文GitPage地址
GitHub: Karobben
Blog:Karobben
BiliBili:史上最不正經的生物狗

