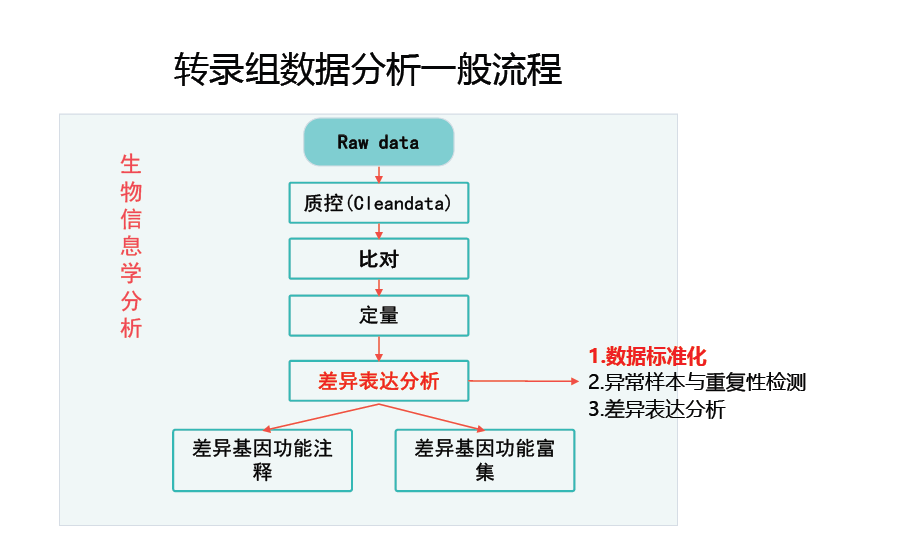
标准化

原因

测序深度
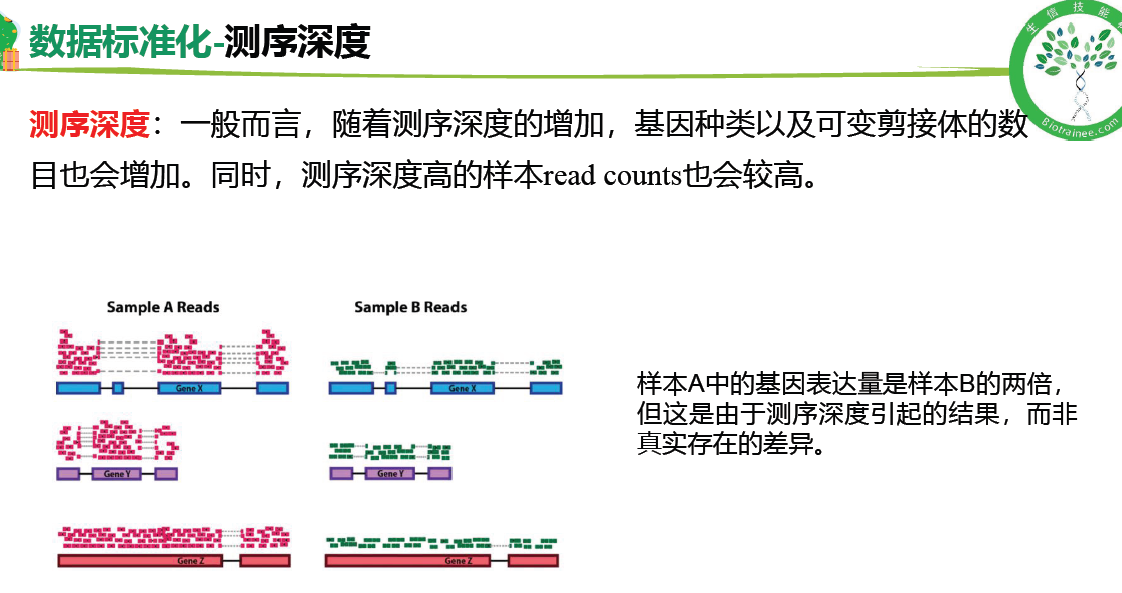
基因长度
FPKM (标化了深度和长度)

RPM (指标化了深度,适用于miRNA)

TPM (推荐)

使用场景

流程
数据预处理(过滤低表达,标准化表达矩阵)

rm(list = ls())options(stringsAsFactors = F)library(stringr)## ====================1.读取数据# 读取raw count表达矩阵rawcount <- read.table("data/raw_counts.txt",row.names = 1, sep = "\t", header = T)colnames(rawcount)# 查看表达谱rawcount[1:4,1:4]# 去除前的基因表达矩阵情况dim(rawcount)# 获取分组信息group <- read.table("data/filereport_read_run_PRJNA229998_tsv.txt", header = T,sep = "\t", quote = "\"")colnames(group)# 提取表达矩阵对应的样本表型信息group <- group[match(colnames(rawcount), group$run_accession), c("run_accession","sample_title")]group # 差异分析方案为:Dex vs untreated group$sample_title <- str_split_fixed(group$sample_title,pattern = "_", n=2)[,2]groupwrite.table(group,file = "data/group.txt",row.names = F,sep = "\t",quote = F) ## =================== 2.表达矩阵预处理 ==========# 过滤低表达基因keep <- rowSums(rawcount>0) >= floor(0.75*ncol(rawcount))table(keep)filter_count <- rawcount[keep,]filter_count[1:4,1:4]dim(filter_count)# 加载edgeR包计算counts per millio(cpm) 表达矩阵library(edgeR)express_cpm <- cpm(filter_count)express_cpm[1:6,1:6]# 保存表达矩阵和分组结果save(filter_count, express_cpm, group, file = "data/Step01-airwayData.Rdata")
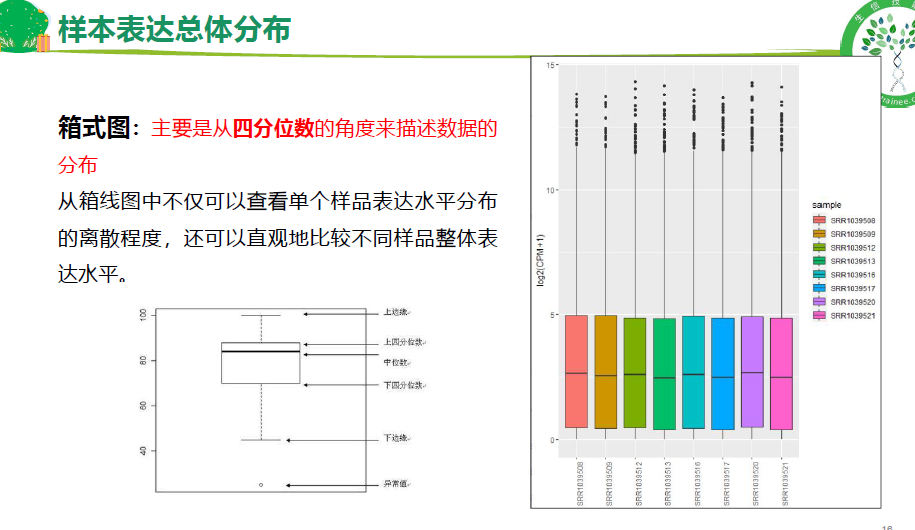
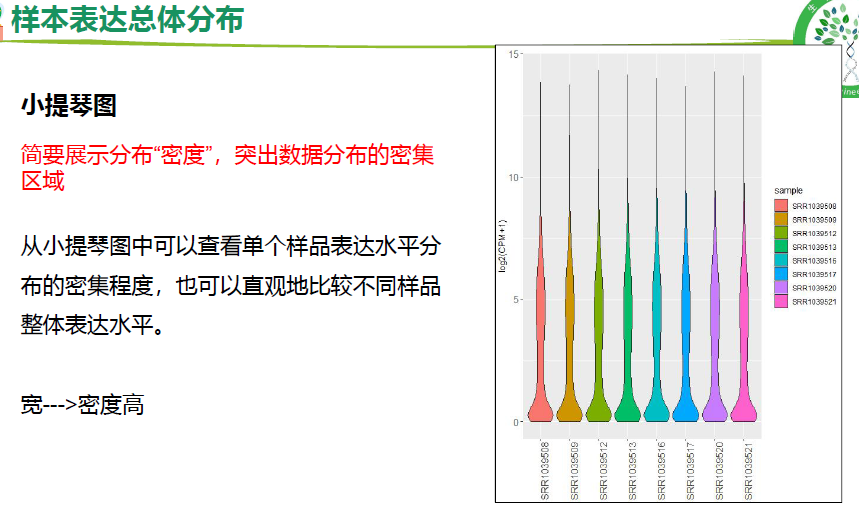
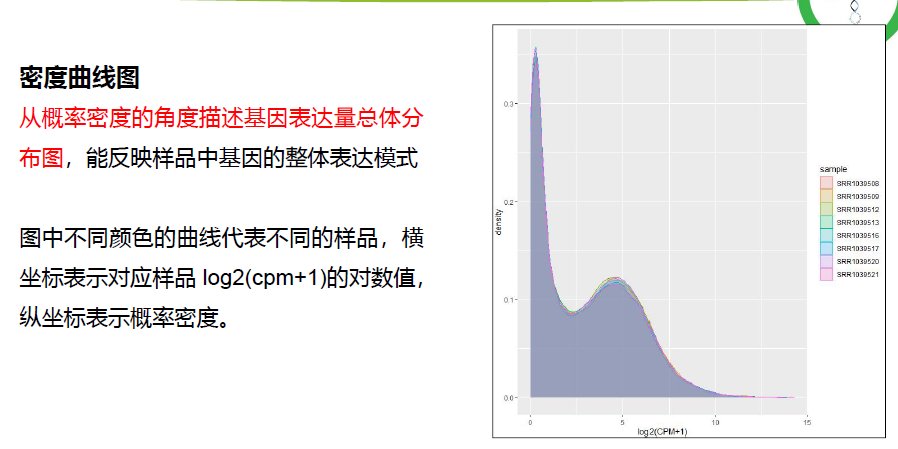
异常和生物学重复检测(箱式图,小提琴图,概率密度分布图)

rm(list = ls())
options(stringsAsFactors = F)
# 加载包,设置绘图参数
library(ggplot2)
library(ggsci)
mythe <- theme_bw() + theme(panel.grid.major=element_blank(),
panel.grid.minor=element_blank())
# 加载原始表达的数据
lname <- load(file = "data/Step01-airwayData.Rdata");lname
exprSet <- log10(as.matrix(express_cpm)+1)
exprSet[1:6,1:6]
## 1.样本表达总体分布-箱式图
# 构造绘图数据
data <- data.frame(expression=c(exprSet),
sample=rep(colnames(exprSet),each=nrow(exprSet)))
head(data)
p <- ggplot(data = data, aes(x=sample,y=expression,fill=sample))
p1 <- p + geom_boxplot() +
mythe+ theme(axis.text.x = element_text(angle = 90)) +
xlab(NULL) + ylab("log10(CPM+1)") ;p1
# 保存图片
ggsave(p1,filename = "result/1.sample_boxplot.pdf",width = 5, height = 5)
## 2.样本表达总体分布-小提琴图
p2 <- p + geom_violin() + mythe +
theme(axis.text = element_text(size = 12),
axis.text.x = element_text(angle = 90)) +
xlab(NULL) + ylab("log10(CPM+1)");p2
# 保存图片
ggsave(p2,filename = "result/1.sample_violin.png", width = 5, height = 5)
## 3.样本表达总体分布-概率密度分布图
m <- ggplot(data=data, aes(x=expression))
p3 <- m + geom_density(aes(fill=sample, colour=sample),alpha = 0.1) +
xlab("log10(CPM+1)") + mythe; p3
# 保存图片
ggsave(p3,filename = "result/1.sample_density.png", width = 5, height = 5)
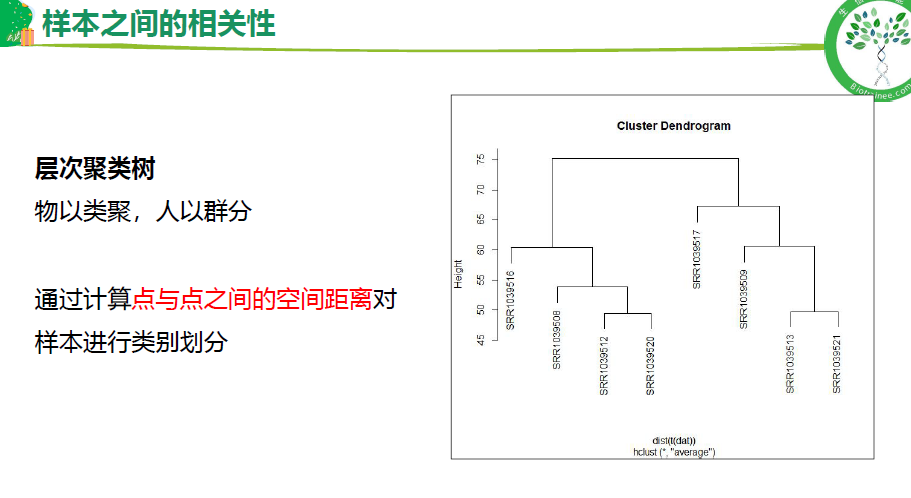
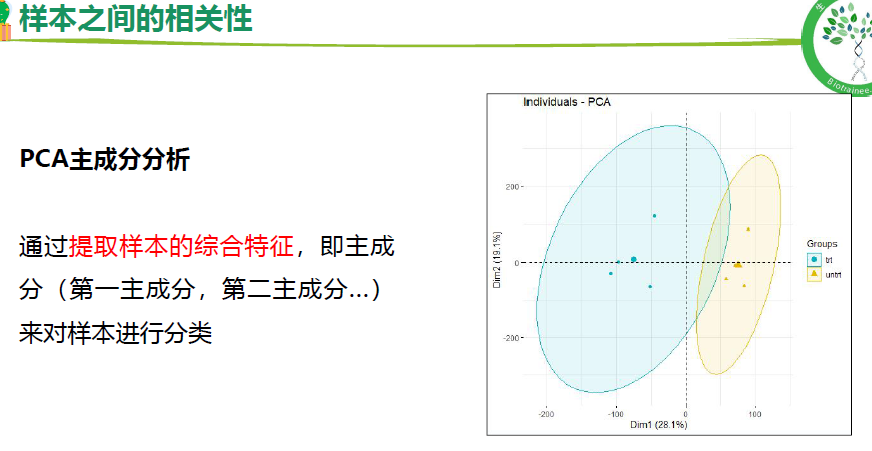
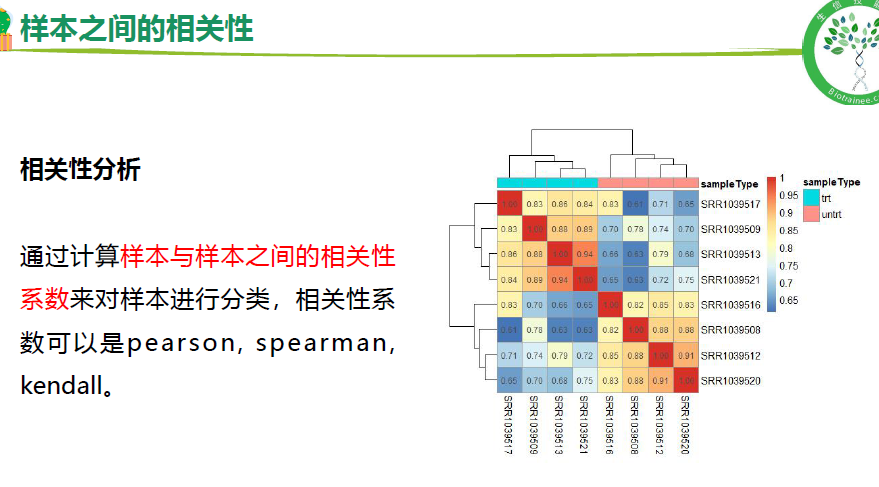
样本相关性(聚类树,PCA, 相关性分析)
# 魔幻操作,一键清空
rm(list = ls())
options(stringsAsFactors = F)
library(FactoMineR)
library(factoextra)
library(corrplot)
library(pheatmap)
library(tidyverse)
# 加载数据并检查
lname <- load(file = 'data/Step01-airwayData.Rdata')
lname
## 1.样本之间的相关性-层次聚类树----
dat <- log10(express_cpm+1)
dat[1:4,1:4]
dim(dat)
sampleTree <- hclust(dist(t(dat)), method = "average")
plot(sampleTree)
# 提取样本聚类信息
temp <- as.data.frame(cutree(sampleTree,k = 2)) %>%
rownames_to_column(var="sample") # 分几类
temp1 <- merge(temp,group,by.x = "sample",by.y="run_accession")
table(temp1$`cutree(sampleTree, k = 2)`,temp1$sample_title) # 表格
# 保存结果
pdf(file = "result/2.sample_Treeplot.pdf",width = 7,height = 6)
plot(sampleTree)
dev.off()
png(file = "result/2.sample_Treeplot.png")
plot(sampleTree)
dev.off()
## 2.样本之间的相关性-PCA----
# 第一步,数据预处理
dat <- log10(express_cpm+1)
dat[1:4,1:4]
dat <- as.data.frame(t(dat))
dat_pca <- PCA(dat, graph = FALSE)
group_list <- group[match(group$run_accession,rownames(dat)), 2];group_list
# geom.ind: point显示点,text显示文字
# palette: 用不同颜色表示分组
# addEllipses: 是否圈起来
mythe <- theme_bw() +
theme(panel.grid.major=element_blank(),panel.grid.minor=element_blank()) +
theme(plot.title = element_text(hjust = 0.5))
p <- fviz_pca_ind(dat_pca,
geom.ind = "text",
col.ind = group_list,
palette = c("#00AFBB", "#E7B800"),
addEllipses = T,
legend.title = "Groups") + mythe;p
# 保存结果
ggsave(p, filename = "result/2.sample_PCA.png",width = 6.5,height = 6)
## 3.样本之间的相关性-cor----
# 选择差异变化大的基因算样本相关性
exprSet <- express_cpm
exprSet = exprSet[names(sort(apply(exprSet, 1, mad),decreasing = T)[1:800]),]
dim(exprSet)
# 计算相关性
M <- cor(exprSet,method = "spearman")
M
# 构造注释条
anno <- data.frame(group=group$sample_title,row.names = group$run_accession )
# 保存结果
pheatmap(M,display_numbers = T,
annotation_col = anno,
fontsize = 10,cellheight = 30,
cellwidth = 30,cluster_rows = T,
cluster_cols = T,
filename = "result/2.sample_Cor.pdf",width = 7.5,height = 7)


















