对于数学建模里用到的多目标优化算法进行探究
使用上次建模选拔赛里做的匆忙且不完整的调参作为优化问题
选择对下面的方程进行拟合确定参数
选择 [1:100] 的100个散点作为拟合区间
def realFuction(x):return -0.02*x**2 +2*x+5*math.sin(x)def myFuc(a1,a2,a3,x):return a1*x**2 + a2*x + a3*math.sin(x)
模拟退火
基于参考链接的代码
但是源代码我认为有一些问题:第57行原代码为r
相对于源代码,我加上了保存最优解的代码,在后面的结果展示里也可以看到对应的结果,防止在最后的遍历里小概率的选择了较差解,导致结果极差
模拟退火参数选择:
- 对于T0决定了一开始参数的变化幅度(参数变化乘了T),所以不易选择过大(容易震荡)或过小
- T0和Tmin又共同决定了温度变化的幅度,影响了参数的变化幅度和退火过程长度
- T = T0 / math.log2(1 + t)决定温度变化速度,不加log更快,但是结果不一定更好
- 对于k即每一个温度下对参数的尝试调整次数 ```python from future import division import numpy as np import matplotlib.pyplot as plt import math from Draw import Draw def realFuction(x): return -0.02x**2 +2x+5*math.sin(x)
def myFuc(a1,a2,a3,x): return a1x**2 + a2x + a3*math.sin(x)
def aimFunction(para,x): res=0 for i in x: res += math.sqrt((myFuc(para[0],para[1],para[2],i)-realFuction(i))**2) return res/len(x)
x = [i for i in range(100)] y = [0 for i in range(100)] for i in range(100): y[i] = realFuction(x[i])
T0 = 200 # initiate temperature\ T=T0 Tmin = 15 # minimum value of terperature
para=[np.random.uniform(low=-1, high=5) for _ in range(3)]
k = 100 # times of internal circulation temp_y = 0 # initiate result t = 0 # time best=[] best_fit=0 while T >= Tmin: for i in range(k):
# calculate ytemp_y = aimFunction(para,x)# generate a new x in the neighboorhood of x by transform functionparaNew = [i+np.random.uniform(low=-0.0005,high=0.0005)*T for i in para]yNew = aimFunction(paraNew,x)if len(best)==0:best_fit=yNewbest=paraNewelse:if yNew<best_fit:best=paraNewbest_fit=yNewif yNew - temp_y < 0:para = paraNewelse:# metropolis principlep = math.exp(-(yNew - temp_y) / T)r = np.random.uniform(low=0, high=1)if r > p:para = paraNewprint(para,aimFunction(para,x),best_fit)t += 1T = T0 / math.log2(1 + t)print("cur T: ",T)
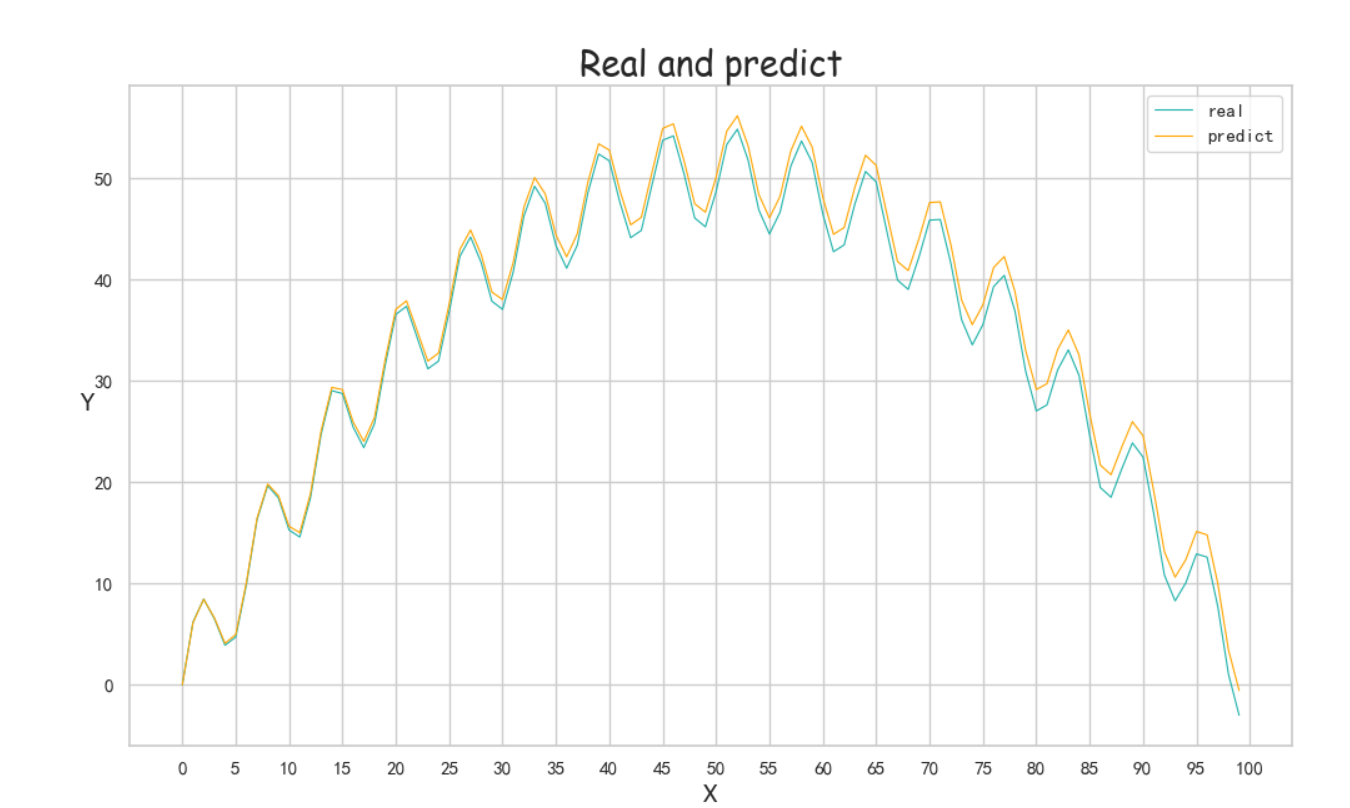
result=[] bestresult=[] for i in x: result.append(myFuc(para[0],para[1],para[2],i)) bestresult.append(myFuc(best[0],best[1],best[2],i)) print(para) print(best) Draw([y,result,bestresult],[“real”,”predict”,”best”],x,”Real and predict”,Colors=None)
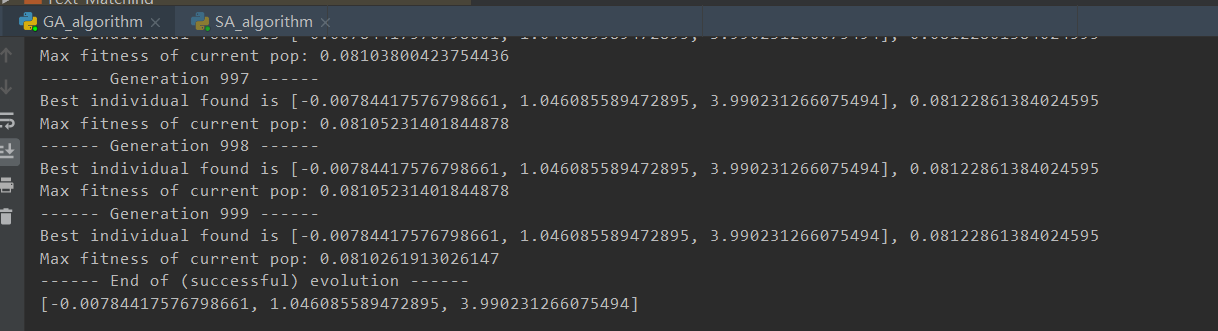
(效果比较好的图忘记截图参数结果了)<br />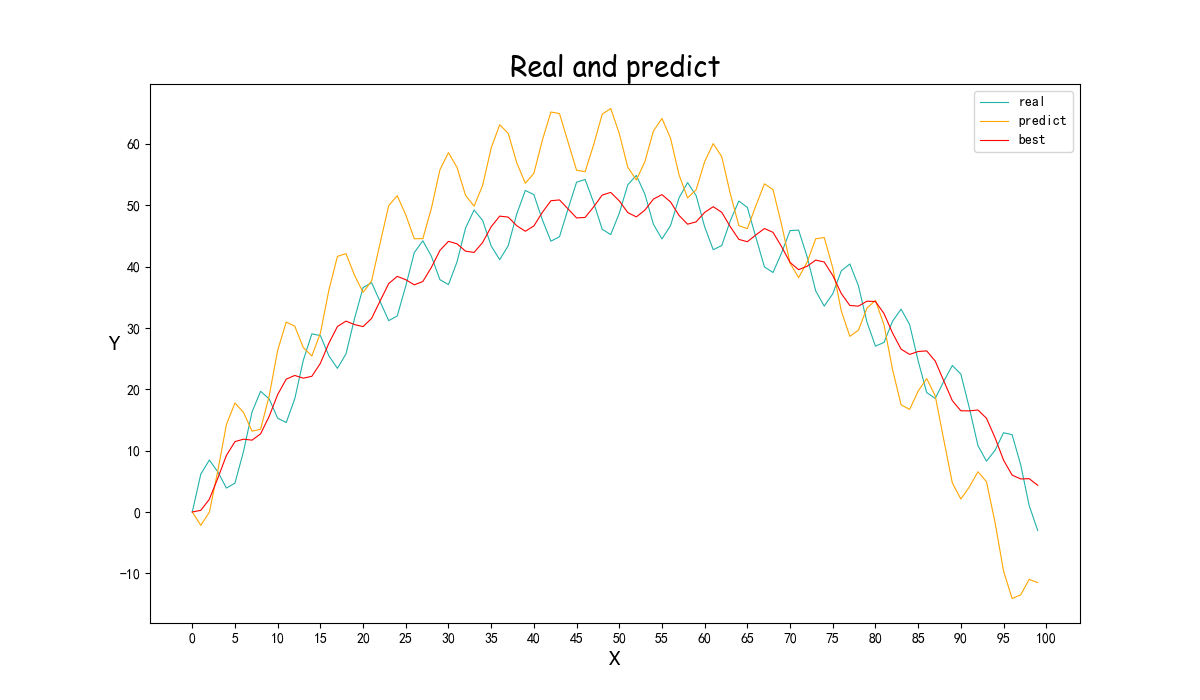<br />(重新跑了,调了一些参数的结果,Best的拟合效果巨好,但是perdict飘了,可以看截图里的最后一次退火计算,不知道为啥第一个参数突变了,但是Best效果实在太好了)<br />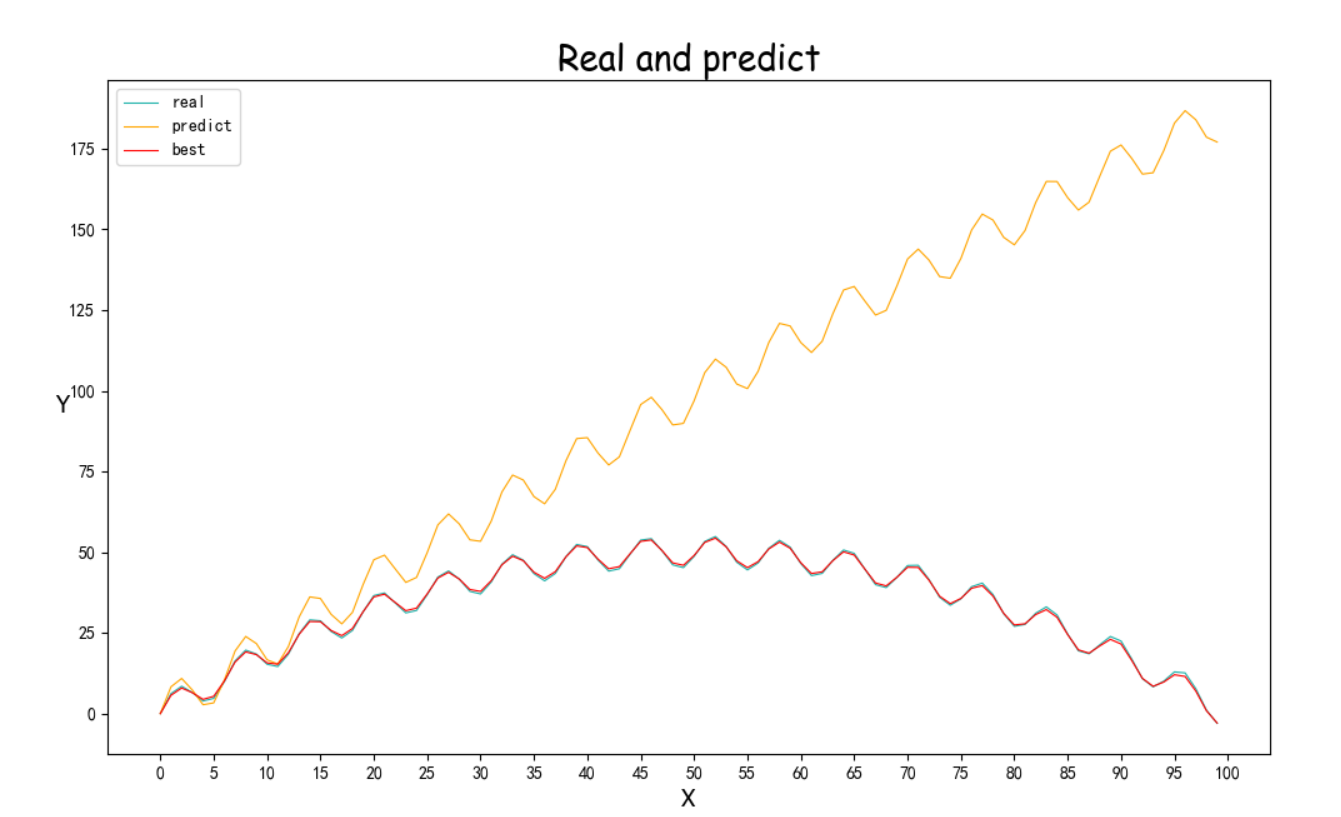<br />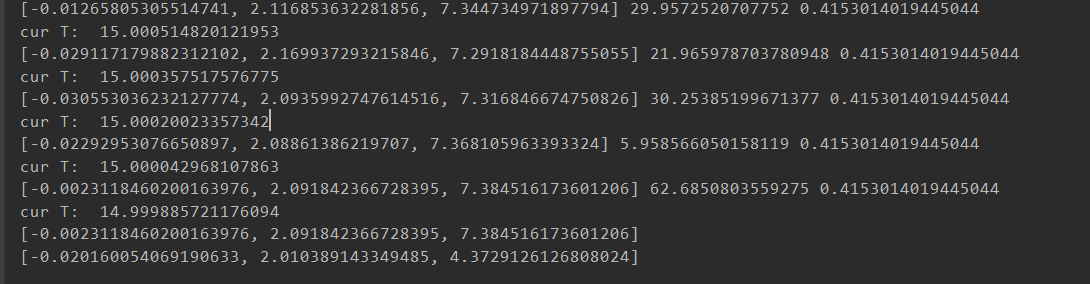<a name="j8Uj0"></a># 遗传算法基于参考资料做了一下修改<br />构造了问题实例和损失函数,同时修改了突变的操作<br />(原来的是在范围里直接随机一个,实在感觉有点随便,对于解空间比较大的参数可能不适合?)<br />最后修改了Fitness的计算<br />原先是fit值向大的方向优化,这里是需要缩小gap<br />第一次改为负号,不可以,会影响到选择的时候(基于Fitness占比选择遗传,负数导致错误)<br />第二次改为倒数```pythonimport randomimport mathimport numpy as npfrom operator import itemgetterdef realFuction(x):return -0.02*x**2 +2*x+5*math.sin(x)def myFuc(a1,a2,a3,x):return a1*x**2 + a2*x + a3*math.sin(x)def aimFunction(para,x):res=0for i in x:res += math.sqrt((myFuc(para[0],para[1],para[2],i)-realFuction(i))**2)return 1/(res/len(x))x = [i for i in range(100)]y = [0 for i in range(100)]for i in range(100):y[i] = realFuction(x[i])class Gene:"""This is a class to represent individual(Gene) in GA algorithomeach object of this class have two attribute: data, size"""def __init__(self, **data):self.__dict__.update(data)self.size = len(data['data']) # length of geneclass GA:"""This is a class of GA algorithm."""def __init__(self, parameter):"""Initialize the pop of GA algorithom and evaluate the pop by computing its' fitness value.The data structure of pop is composed of several individuals which has the form like that:{'Gene':a object of class Gene, 'fitness': 1.02(for example)}Representation of Gene is a list: [b s0 u0 sita0 s1 u1 sita1 s2 u2 sita2]"""# parameter = [CXPB, MUTPB, NGEN, popsize, low, up]self.parameter = parameterlow = self.parameter[4]up = self.parameter[5]self.bound = []self.bound.append(low)self.bound.append(up)pop = []for i in range(self.parameter[3]):geneinfo = []for pos in range(len(low)):geneinfo.append(random.randint(self.bound[0][pos], self.bound[1][pos])) # initialise popluationfitness = self.evaluate(geneinfo) # evaluate each chromosomepop.append({'Gene': Gene(data=geneinfo), 'fitness': fitness}) # store the chromosome and its fitnessself.pop = popself.bestindividual = self.selectBest(self.pop) # store the best chromosome in the populationdef evaluate(self, geneinfo):"""fitness function"""y = aimFunction(geneinfo,x)return ydef selectBest(self, pop):"""select the best individual from pop"""s_inds = sorted(pop, key=itemgetter("fitness"), reverse=True) # from large to small, return a popreturn s_inds[0]def selection(self, individuals, k):"""select some good individuals from pop, note that good individuals have greater probability to be choosenfor example: a fitness list like that:[5, 4, 3, 2, 1], sum is 15,[-----|----|---|--|-]012345|6789|101112|1314|15we randomly choose a value in [0, 15],it belongs to first scale with greatest probability"""s_inds = sorted(individuals, key=itemgetter("fitness"),reverse=True) # sort the pop by the reference of fitnesssum_fits = sum(ind['fitness'] for ind in individuals) # sum up the fitness of the whole popchosen = []for i in range(k):u = random.random() * sum_fits # randomly produce a num in the range of [0, sum_fits], as thresholdsum_ = 0for ind in s_inds:sum_ += ind['fitness'] # sum up the fitnessif sum_ >= u:# when the sum of fitness is bigger than u, choose the one, which means u is in the range of# [sum(1,2,...,n-1),sum(1,2,...,n)] and is time to choose the one ,namely n-th individual in the popchosen.append(ind)break# from small to large, due to list.pop() method get the last elementchosen = sorted(chosen, key=itemgetter("fitness"), reverse=False)return chosendef crossoperate(self, offspring):"""cross operationhere we use two points crossoperatefor example: gene1: [5, 2, 4, 7], gene2: [3, 6, 9, 2], if pos1=1, pos2=25 | 2 | 4 73 | 6 | 9 2=3 | 2 | 9 25 | 6 | 4 7"""dim = len(offspring[0]['Gene'].data)geninfo1 = offspring[0]['Gene'].data # Gene's data of first offspring chosen from the selected popgeninfo2 = offspring[1]['Gene'].data # Gene's data of second offspring chosen from the selected popif dim == 1:pos1 = 1pos2 = 1else:pos1 = random.randrange(1, dim) # select a position in the range from 0 to dim-1,pos2 = random.randrange(1, dim)newoff1 = Gene(data=[]) # offspring1 produced by cross operationnewoff2 = Gene(data=[]) # offspring2 produced by cross operationtemp1 = []temp2 = []for i in range(dim):if min(pos1, pos2) <= i < max(pos1, pos2):temp2.append(geninfo2[i])temp1.append(geninfo1[i])else:temp2.append(geninfo1[i])temp1.append(geninfo2[i])newoff1.data = temp1newoff2.data = temp2return newoff1, newoff2def mutation(self, crossoff, bound):"""mutation operation"""'''dim = len(crossoff.data)if dim == 1:pos = 0else:pos = random.randrange(0, dim) # chose a position in crossoff to perform mutation.#crossoff.data[pos] = random.uniform(bound[0][pos], bound[1][pos])'''crossoff.data = [i + np.random.uniform(low=-0.001, high=0.001) for i in crossoff.data]return crossoffdef GA_main(self):"""main frame work of GA"""popsize = self.parameter[3]print("Start of evolution")# Begin the evolutionfor g in range(NGEN):print("------ Generation {} ------".format(g))# Apply selection based on their converted fitnessselectpop = self.selection(self.pop, popsize)nextoff = []while len(nextoff) != popsize:# Apply crossover and mutation on the offspring# Select two individualsoffspring = [selectpop.pop() for _ in range(2)]if random.random() < CXPB: # cross two individuals with probability CXPBcrossoff1, crossoff2 = self.crossoperate(offspring)if random.random() < MUTPB: # mutate an individual with probability MUTPBmuteoff1 = self.mutation(crossoff1, self.bound)muteoff2 = self.mutation(crossoff2, self.bound)fit_muteoff1 = self.evaluate(muteoff1.data) # Evaluate the individualsfit_muteoff2 = self.evaluate(muteoff2.data) # Evaluate the individualsnextoff.append({'Gene': muteoff1, 'fitness': fit_muteoff1})nextoff.append({'Gene': muteoff2, 'fitness': fit_muteoff2})else:fit_crossoff1 = self.evaluate(crossoff1.data) # Evaluate the individualsfit_crossoff2 = self.evaluate(crossoff2.data)nextoff.append({'Gene': crossoff1, 'fitness': fit_crossoff1})nextoff.append({'Gene': crossoff2, 'fitness': fit_crossoff2})else:nextoff.extend(offspring)# The population is entirely replaced by the offspringself.pop = nextoff# Gather all the fitnesses in one list and print the statsfits = [ind['fitness'] for ind in self.pop]best_ind = self.selectBest(self.pop)if best_ind['fitness'] > self.bestindividual['fitness']:self.bestindividual = best_indprint("Best individual found is {}, {}".format(self.bestindividual['Gene'].data,self.bestindividual['fitness']))print("Max fitness of current pop: {}".format(max(fits)))print("------ End of (successful) evolution ------")from Draw import Drawif __name__ == "__main__":CXPB, MUTPB, NGEN, popsize = 0.7, 0.5, 1000, 1000#control parametersup = [5, 5, 5] # upper range for variableslow = [-2, -2, -2] # lower range for variablesparameter = [CXPB, MUTPB, NGEN, popsize, low, up]run = GA(parameter)run.GA_main()print(run.bestindividual['Gene'].data)result = []for i in x:result.append(myFuc(run.bestindividual['Gene'].data[0], run.bestindividual['Gene'].data[1], run.bestindividual['Gene'].data[2], i))Draw([y, result], ["real", "predict"], x, "Real and predict", Colors=None)
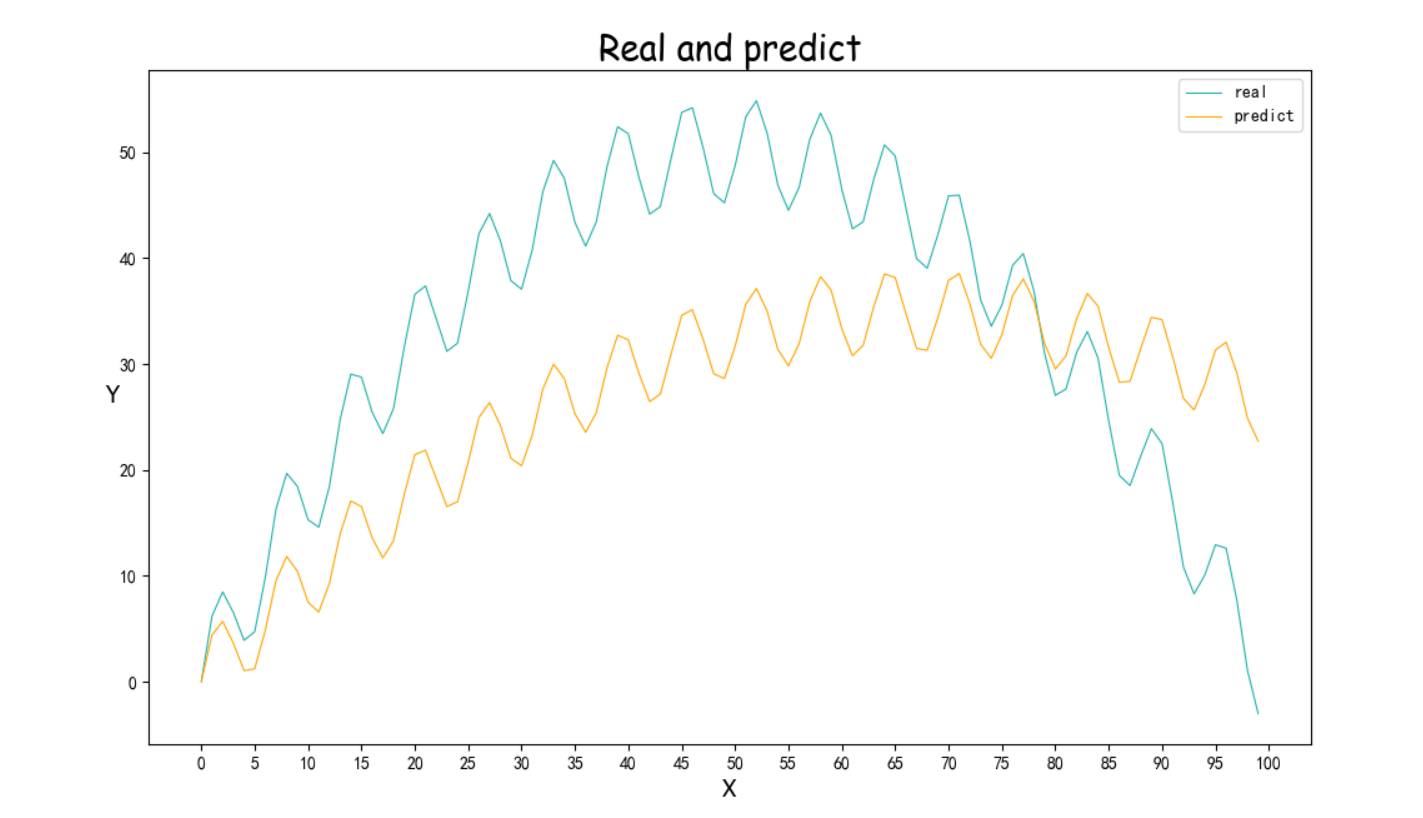
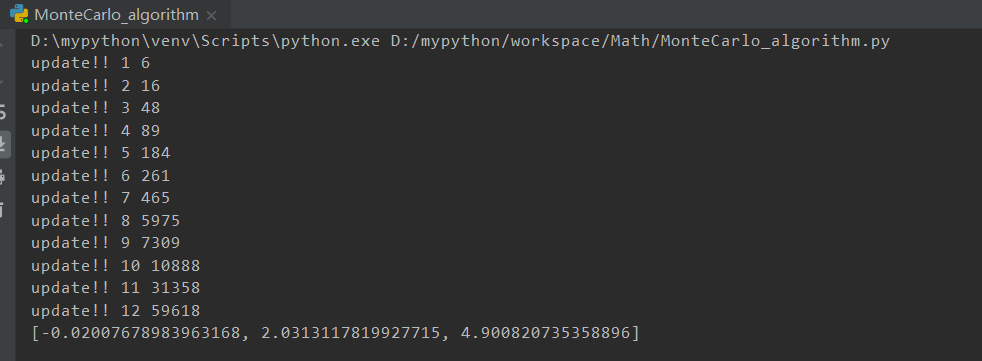
蒙特卡洛算法
还有一种算法就是使用蒙特卡洛进行优化
优化的方法很简单,就是直接随机的生成参数,然后进行尝试,如果优化则保留
简单的基于模拟退火的算法框架实现如下
from __future__ import divisionimport numpy as npimport matplotlib.pyplot as pltimport mathfrom Draw import Drawdef realFuction(x):return -0.02*x**2 +2*x+5*math.sin(x)def myFuc(a1,a2,a3,x):return a1*x**2 + a2*x + a3*math.sin(x)def aimFunction(para,x):res=0for i in x:res += math.sqrt((myFuc(para[0],para[1],para[2],i)-realFuction(i))**2)return res/len(x)x = [i for i in range(100)]y = [0 for i in range(100)]for i in range(100):y[i] = realFuction(x[i])Need=[np.random.uniform(low=-1, high=5) for _ in range(3)]cur_best=aimFunction(Need,x)update_time=0for i in range(100000):para=[np.random.uniform(low=-1, high=5) for _ in range(3)]temp_y = aimFunction(para,x)if temp_y - cur_best < 0:Need=paracur_best=temp_yupdate_time+=1print("update!!",update_time,i)result=[]print(Need)for i in x:result.append(myFuc(Need[0],Need[1],Need[2],i))Draw([y,result],["real","predict"],x,"Real and predict",Colors=None)
备注
绘图函数,基于数学建模选拔赛代码整合封装得到
import numpy as npimport matplotlib.pyplot as pltfrom matplotlib.pyplot import MultipleLocatorplt.rcParams['font.sans-serif']=['SimHei'] #用来正常显示中文标签plt.rcParams['axes.unicode_minus']=False #用来正常显示负号#families=[ 'fantasy','Tahoma', 'monospace','Times New Roman']def Draw(Lines,Names,index,Title,Colors=None):if Colors==None:Colors=['lightseagreen','orange','red','darkblue','green', ]plt.rcParams['figure.figsize'] = (12.0, 7.0)for i in range(len(Lines)):plt.plot(index,Lines[i],Colors[i],label=Names[i],linewidth=0.8)Title_font = {'family': 'fantasy','size': 20}plt.title(Title, Title_font)font1 = {'family': 'Tahoma','size': 14}plt.xlabel('X', font1)plt.ylabel('Y', font1,rotation=0)#plt.ylabel('Y', font1)plt.legend()x_len=len(index)/20x_major_locator = MultipleLocator(x_len)ax = plt.gca()ax.xaxis.set_major_locator(x_major_locator)plt.show()#X = np.linspace(-np.pi, np.pi, 256, endpoint=True)#C,S = np.cos(X), np.sin(X)#Draw([C,S],['Sin','Cos'],X,"test")

