- Indexing and selecting data
- Different choices for indexing
- Basics
- Attribute access
- Slicing ranges
- Selection by label
- Selection by position
- Selection by callable
- IX indexer is deprecated
- Indexing with list with missing labels is deprecated
- Selecting random samples
- Setting with enlargement
- Fast scalar value getting and setting
- Boolean indexing
- Indexing with isin
- The
where()Method and Masking - The
query()Method - Duplicate data
- Dictionary-like
get()method - The
lookup()method - Index objects
- Set / reset index
- Returning a view versus a copy
Indexing and selecting data
The axis labeling information in pandas objects serves many purposes:
- Identifies data (i.e. provides metadata) using known indicators, important for analysis, visualization, and interactive console display.
- Enables automatic and explicit data alignment.
- Allows intuitive getting and setting of subsets of the data set.
In this section, we will focus on the final point: namely, how to slice, dice, and generally get and set subsets of pandas objects. The primary focus will be on Series and DataFrame as they have received more development attention in this area.
::: tip Note
The Python and NumPy indexing operators [] and attribute operator .
provide quick and easy access to pandas data structures across a wide range
of use cases. This makes interactive work intuitive, as there’s little new
to learn if you already know how to deal with Python dictionaries and NumPy
arrays. However, since the type of the data to be accessed isn’t known in
advance, directly using standard operators has some optimization limits. For
production code, we recommended that you take advantage of the optimized
pandas data access methods exposed in this chapter.
:::
::: danger Warning
Whether a copy or a reference is returned for a setting operation, may
depend on the context. This is sometimes called chained assignment and
should be avoided. See Returning a View versus Copy.
:::
::: danger Warning
Indexing on an integer-based Index with floats has been clarified in 0.18.0, for a summary of the changes, see here.
:::
See the MultiIndex / Advanced Indexing for MultiIndex and more advanced indexing documentation.
See the cookbook for some advanced strategies.
Different choices for indexing
Object selection has had a number of user-requested additions in order to support more explicit location based indexing. Pandas now supports three types of multi-axis indexing.
.locis primarily label based, but may also be used with a boolean array..locwill raiseKeyErrorwhen the items are not found. Allowed inputs are:- A single label, e.g.
5or'a'(Note that5is interpreted as a label of the index. This use is not an integer position along the index.). - A list or array of labels
['a', 'b', 'c']. - A slice object with labels
'a':'f'(Note that contrary to usual python slices, both the start and the stop are included, when present in the index! See Slicing with labels and Endpoints are inclusive.) - A boolean array
- A
callablefunction with one argument (the calling Series or DataFrame) and that returns valid output for indexing (one of the above).
New in version 0.18.1.
See more at Selection by Label.
- A single label, e.g.
.ilocis primarily integer position based (from0tolength-1of the axis), but may also be used with a boolean array..ilocwill raiseIndexErrorif a requested indexer is out-of-bounds, except slice indexers which allow out-of-bounds indexing. (this conforms with Python/NumPy slice semantics). Allowed inputs are:- An integer e.g.
5. - A list or array of integers
[4, 3, 0]. - A slice object with ints
1:7. - A boolean array.
- A
callablefunction with one argument (the calling Series or DataFrame) and that returns valid output for indexing (one of the above).
New in version 0.18.1.
See more at Selection by Position, Advanced Indexing and Advanced Hierarchical.
- An integer e.g.
5. - A list or array of integers
[4, 3, 0]. - A slice object with ints
1:7. - A boolean array.
- A
callablefunction with one argument (the calling Series or DataFrame) and that returns valid output for indexing (one of the above).
New in version 0.18.1.
- An integer e.g.
.loc,.iloc, and also[]indexing can accept acallableas indexer. See more at Selection By Callable.
Getting values from an object with multi-axes selection uses the following
notation (using .loc as an example, but the following applies to .iloc as
well). Any of the axes accessors may be the null slice :. Axes left out of
the specification are assumed to be :, e.g. p.loc['a'] is equivalent to
p.loc['a', :, :].
| Object Type | Indexers |
|---|---|
| Series | s.loc[indexer] |
| DataFrame | df.loc[row_indexer,column_indexer] |
Basics
As mentioned when introducing the data structures in the last section, the primary function of indexing with [] (a.k.a. __getitem__
for those familiar with implementing class behavior in Python) is selecting out
lower-dimensional slices. The following table shows return type values when
indexing pandas objects with []:
| Object Type | Selection | Return Value Type |
|---|---|---|
| Series | series[label] | scalar value |
| DataFrame | frame[colname] | Series corresponding to colname |
Here we construct a simple time series data set to use for illustrating the indexing functionality:
In [1]: dates = pd.date_range('1/1/2000', periods=8)In [2]: df = pd.DataFrame(np.random.randn(8, 4),...: index=dates, columns=['A', 'B', 'C', 'D'])...:In [3]: dfOut[3]:A B C D2000-01-01 0.469112 -0.282863 -1.509059 -1.1356322000-01-02 1.212112 -0.173215 0.119209 -1.0442362000-01-03 -0.861849 -2.104569 -0.494929 1.0718042000-01-04 0.721555 -0.706771 -1.039575 0.2718602000-01-05 -0.424972 0.567020 0.276232 -1.0874012000-01-06 -0.673690 0.113648 -1.478427 0.5249882000-01-07 0.404705 0.577046 -1.715002 -1.0392682000-01-08 -0.370647 -1.157892 -1.344312 0.844885
::: tip Note
None of the indexing functionality is time series specific unless specifically stated.
:::
Thus, as per above, we have the most basic indexing using []:
In [4]: s = df['A']In [5]: s[dates[5]]Out[5]: -0.6736897080883706
You can pass a list of columns to [] to select columns in that order.
If a column is not contained in the DataFrame, an exception will be
raised. Multiple columns can also be set in this manner:
In [6]: dfOut[6]:A B C D2000-01-01 0.469112 -0.282863 -1.509059 -1.1356322000-01-02 1.212112 -0.173215 0.119209 -1.0442362000-01-03 -0.861849 -2.104569 -0.494929 1.0718042000-01-04 0.721555 -0.706771 -1.039575 0.2718602000-01-05 -0.424972 0.567020 0.276232 -1.0874012000-01-06 -0.673690 0.113648 -1.478427 0.5249882000-01-07 0.404705 0.577046 -1.715002 -1.0392682000-01-08 -0.370647 -1.157892 -1.344312 0.844885In [7]: df[['B', 'A']] = df[['A', 'B']]In [8]: dfOut[8]:A B C D2000-01-01 -0.282863 0.469112 -1.509059 -1.1356322000-01-02 -0.173215 1.212112 0.119209 -1.0442362000-01-03 -2.104569 -0.861849 -0.494929 1.0718042000-01-04 -0.706771 0.721555 -1.039575 0.2718602000-01-05 0.567020 -0.424972 0.276232 -1.0874012000-01-06 0.113648 -0.673690 -1.478427 0.5249882000-01-07 0.577046 0.404705 -1.715002 -1.0392682000-01-08 -1.157892 -0.370647 -1.344312 0.844885
You may find this useful for applying a transform (in-place) to a subset of the columns.
::: danger Warning
pandas aligns all AXES when setting Series and DataFrame from .loc, and .iloc.
This will not modify df because the column alignment is before value assignment.
In [9]: df[['A', 'B']]Out[9]:A B2000-01-01 -0.282863 0.4691122000-01-02 -0.173215 1.2121122000-01-03 -2.104569 -0.8618492000-01-04 -0.706771 0.7215552000-01-05 0.567020 -0.4249722000-01-06 0.113648 -0.6736902000-01-07 0.577046 0.4047052000-01-08 -1.157892 -0.370647In [10]: df.loc[:, ['B', 'A']] = df[['A', 'B']]In [11]: df[['A', 'B']]Out[11]:A B2000-01-01 -0.282863 0.4691122000-01-02 -0.173215 1.2121122000-01-03 -2.104569 -0.8618492000-01-04 -0.706771 0.7215552000-01-05 0.567020 -0.4249722000-01-06 0.113648 -0.6736902000-01-07 0.577046 0.4047052000-01-08 -1.157892 -0.370647
The correct way to swap column values is by using raw values:
In [12]: df.loc[:, ['B', 'A']] = df[['A', 'B']].to_numpy()In [13]: df[['A', 'B']]Out[13]:A B2000-01-01 0.469112 -0.2828632000-01-02 1.212112 -0.1732152000-01-03 -0.861849 -2.1045692000-01-04 0.721555 -0.7067712000-01-05 -0.424972 0.5670202000-01-06 -0.673690 0.1136482000-01-07 0.404705 0.5770462000-01-08 -0.370647 -1.157892
:::
Attribute access
You may access an index on a Series or column on a DataFrame directly
as an attribute:
In [14]: sa = pd.Series([1, 2, 3], index=list('abc'))In [15]: dfa = df.copy()
In [16]: sa.bOut[16]: 2In [17]: dfa.AOut[17]:2000-01-01 0.4691122000-01-02 1.2121122000-01-03 -0.8618492000-01-04 0.7215552000-01-05 -0.4249722000-01-06 -0.6736902000-01-07 0.4047052000-01-08 -0.370647Freq: D, Name: A, dtype: float64
In [18]: sa.a = 5In [19]: saOut[19]:a 5b 2c 3dtype: int64In [20]: dfa.A = list(range(len(dfa.index))) # ok if A already existsIn [21]: dfaOut[21]:A B C D2000-01-01 0 -0.282863 -1.509059 -1.1356322000-01-02 1 -0.173215 0.119209 -1.0442362000-01-03 2 -2.104569 -0.494929 1.0718042000-01-04 3 -0.706771 -1.039575 0.2718602000-01-05 4 0.567020 0.276232 -1.0874012000-01-06 5 0.113648 -1.478427 0.5249882000-01-07 6 0.577046 -1.715002 -1.0392682000-01-08 7 -1.157892 -1.344312 0.844885In [22]: dfa['A'] = list(range(len(dfa.index))) # use this form to create a new columnIn [23]: dfaOut[23]:A B C D2000-01-01 0 -0.282863 -1.509059 -1.1356322000-01-02 1 -0.173215 0.119209 -1.0442362000-01-03 2 -2.104569 -0.494929 1.0718042000-01-04 3 -0.706771 -1.039575 0.2718602000-01-05 4 0.567020 0.276232 -1.0874012000-01-06 5 0.113648 -1.478427 0.5249882000-01-07 6 0.577046 -1.715002 -1.0392682000-01-08 7 -1.157892 -1.344312 0.844885
::: danger Warning
- You can use this access only if the index element is a valid Python identifier, e.g.
s.1is not allowed. See here for an explanation of valid identifiers. - The attribute will not be available if it conflicts with an existing method name, e.g.
s.minis not allowed. - Similarly, the attribute will not be available if it conflicts with any of the following list:
index,major_axis,minor_axis,items. - In any of these cases, standard indexing will still work, e.g.
s['1'],s['min'], ands['index']will access the corresponding element or column.
:::
If you are using the IPython environment, you may also use tab-completion to see these accessible attributes.
You can also assign a dict to a row of a DataFrame:
In [24]: x = pd.DataFrame({'x': [1, 2, 3], 'y': [3, 4, 5]})In [25]: x.iloc[1] = {'x': 9, 'y': 99}In [26]: xOut[26]:x y0 1 31 9 992 3 5
You can use attribute access to modify an existing element of a Series or column of a DataFrame, but be careful;
if you try to use attribute access to create a new column, it creates a new attribute rather than a
new column. In 0.21.0 and later, this will raise a UserWarning:
In [1]: df = pd.DataFrame({'one': [1., 2., 3.]})In [2]: df.two = [4, 5, 6]UserWarning: Pandas doesn't allow Series to be assigned into nonexistent columns - see https://pandas.pydata.org/pandas-docs/stable/indexing.html#attribute_accessIn [3]: dfOut[3]:one0 1.01 2.02 3.0
Slicing ranges
The most robust and consistent way of slicing ranges along arbitrary axes is
described in the Selection by Position section
detailing the .iloc method. For now, we explain the semantics of slicing using the [] operator.
With Series, the syntax works exactly as with an ndarray, returning a slice of the values and the corresponding labels:
In [27]: s[:5]Out[27]:2000-01-01 0.4691122000-01-02 1.2121122000-01-03 -0.8618492000-01-04 0.7215552000-01-05 -0.424972Freq: D, Name: A, dtype: float64In [28]: s[::2]Out[28]:2000-01-01 0.4691122000-01-03 -0.8618492000-01-05 -0.4249722000-01-07 0.404705Freq: 2D, Name: A, dtype: float64In [29]: s[::-1]Out[29]:2000-01-08 -0.3706472000-01-07 0.4047052000-01-06 -0.6736902000-01-05 -0.4249722000-01-04 0.7215552000-01-03 -0.8618492000-01-02 1.2121122000-01-01 0.469112Freq: -1D, Name: A, dtype: float64
Note that setting works as well:
In [30]: s2 = s.copy()In [31]: s2[:5] = 0In [32]: s2Out[32]:2000-01-01 0.0000002000-01-02 0.0000002000-01-03 0.0000002000-01-04 0.0000002000-01-05 0.0000002000-01-06 -0.6736902000-01-07 0.4047052000-01-08 -0.370647Freq: D, Name: A, dtype: float64
With DataFrame, slicing inside of [] slices the rows. This is provided
largely as a convenience since it is such a common operation.
In [33]: df[:3]Out[33]:A B C D2000-01-01 0.469112 -0.282863 -1.509059 -1.1356322000-01-02 1.212112 -0.173215 0.119209 -1.0442362000-01-03 -0.861849 -2.104569 -0.494929 1.071804In [34]: df[::-1]Out[34]:A B C D2000-01-08 -0.370647 -1.157892 -1.344312 0.8448852000-01-07 0.404705 0.577046 -1.715002 -1.0392682000-01-06 -0.673690 0.113648 -1.478427 0.5249882000-01-05 -0.424972 0.567020 0.276232 -1.0874012000-01-04 0.721555 -0.706771 -1.039575 0.2718602000-01-03 -0.861849 -2.104569 -0.494929 1.0718042000-01-02 1.212112 -0.173215 0.119209 -1.0442362000-01-01 0.469112 -0.282863 -1.509059 -1.135632
Selection by label
::: danger Warning
Whether a copy or a reference is returned for a setting operation, may depend on the context.
This is sometimes called chained assignment and should be avoided.
See Returning a View versus Copy.
:::
::: danger Warning
In [35]: dfl = pd.DataFrame(np.random.randn(5, 4),....: columns=list('ABCD'),....: index=pd.date_range('20130101', periods=5))....:In [36]: dflOut[36]:A B C D2013-01-01 1.075770 -0.109050 1.643563 -1.4693882013-01-02 0.357021 -0.674600 -1.776904 -0.9689142013-01-03 -1.294524 0.413738 0.276662 -0.4720352013-01-04 -0.013960 -0.362543 -0.006154 -0.9230612013-01-05 0.895717 0.805244 -1.206412 2.565646
In [4]: dfl.loc[2:3]TypeError: cannot do slice indexing on <class 'pandas.tseries.index.DatetimeIndex'> with these indexers [2] of <type 'int'>
String likes in slicing can be convertible to the type of the index and lead to natural slicing.
In [37]: dfl.loc['20130102':'20130104']Out[37]:A B C D2013-01-02 0.357021 -0.674600 -1.776904 -0.9689142013-01-03 -1.294524 0.413738 0.276662 -0.4720352013-01-04 -0.013960 -0.362543 -0.006154 -0.923061
:::
::: danger Warning
Starting in 0.21.0, pandas will show a FutureWarning if indexing with a list with missing labels. In the future
this will raise a KeyError. See list-like Using loc with missing keys in a list is Deprecated.
:::
pandas provides a suite of methods in order to have purely label based indexing. This is a strict inclusion based protocol.
Every label asked for must be in the index, or a KeyError will be raised.
When slicing, both the start bound AND the stop bound are included, if present in the index.
Integers are valid labels, but they refer to the label and not the position.
The .loc attribute is the primary access method. The following are valid inputs:
- A single label, e.g.
5or'a'(Note that5is interpreted as a label of the index. This use is not an integer position along the index.). - A list or array of labels
['a', 'b', 'c']. - A slice object with labels
'a':'f'(Note that contrary to usual python slices, both the start and the stop are included, when present in the index! See Slicing with labels. - A boolean array.
- A
callable, see Selection By Callable.
In [38]: s1 = pd.Series(np.random.randn(6), index=list('abcdef'))In [39]: s1Out[39]:a 1.431256b 1.340309c -1.170299d -0.226169e 0.410835f 0.813850dtype: float64In [40]: s1.loc['c':]Out[40]:c -1.170299d -0.226169e 0.410835f 0.813850dtype: float64In [41]: s1.loc['b']Out[41]: 1.3403088497993827
Note that setting works as well:
In [42]: s1.loc['c':] = 0In [43]: s1Out[43]:a 1.431256b 1.340309c 0.000000d 0.000000e 0.000000f 0.000000dtype: float64
With a DataFrame:
In [44]: df1 = pd.DataFrame(np.random.randn(6, 4),....: index=list('abcdef'),....: columns=list('ABCD'))....:In [45]: df1Out[45]:A B C Da 0.132003 -0.827317 -0.076467 -1.187678b 1.130127 -1.436737 -1.413681 1.607920c 1.024180 0.569605 0.875906 -2.211372d 0.974466 -2.006747 -0.410001 -0.078638e 0.545952 -1.219217 -1.226825 0.769804f -1.281247 -0.727707 -0.121306 -0.097883In [46]: df1.loc[['a', 'b', 'd'], :]Out[46]:A B C Da 0.132003 -0.827317 -0.076467 -1.187678b 1.130127 -1.436737 -1.413681 1.607920d 0.974466 -2.006747 -0.410001 -0.078638
Accessing via label slices:
In [47]: df1.loc['d':, 'A':'C']Out[47]:A B Cd 0.974466 -2.006747 -0.410001e 0.545952 -1.219217 -1.226825f -1.281247 -0.727707 -0.121306
For getting a cross section using a label (equivalent to df.xs('a')):
In [48]: df1.loc['a']Out[48]:A 0.132003B -0.827317C -0.076467D -1.187678Name: a, dtype: float64
For getting values with a boolean array:
In [49]: df1.loc['a'] > 0Out[49]:A TrueB FalseC FalseD FalseName: a, dtype: boolIn [50]: df1.loc[:, df1.loc['a'] > 0]Out[50]:Aa 0.132003b 1.130127c 1.024180d 0.974466e 0.545952f -1.281247
For getting a value explicitly (equivalent to deprecated df.get_value('a','A')):
# this is also equivalent to ``df1.at['a','A']``In [51]: df1.loc['a', 'A']Out[51]: 0.13200317033032932
Slicing with labels
When using .loc with slices, if both the start and the stop labels are
present in the index, then elements located between the two (including them)
are returned:
In [52]: s = pd.Series(list('abcde'), index=[0, 3, 2, 5, 4])In [53]: s.loc[3:5]Out[53]:3 b2 c5 ddtype: object
If at least one of the two is absent, but the index is sorted, and can be compared against start and stop labels, then slicing will still work as expected, by selecting labels which rank between the two:
In [54]: s.sort_index()Out[54]:0 a2 c3 b4 e5 ddtype: objectIn [55]: s.sort_index().loc[1:6]Out[55]:2 c3 b4 e5 ddtype: object
However, if at least one of the two is absent and the index is not sorted, an
error will be raised (since doing otherwise would be computationally expensive,
as well as potentially ambiguous for mixed type indexes). For instance, in the
above example, s.loc[1:6] would raise KeyError.
For the rationale behind this behavior, see Endpoints are inclusive.
Selection by position
::: danger Warning
Whether a copy or a reference is returned for a setting operation, may depend on the context.
This is sometimes called chained assignment and should be avoided.
See Returning a View versus Copy.
:::
Pandas provides a suite of methods in order to get purely integer based indexing. The semantics follow closely Python and NumPy slicing. These are 0-based indexing. When slicing, the start bound is included, while the upper bound is excluded. Trying to use a non-integer, even a valid label will raise an IndexError.
The .iloc attribute is the primary access method. The following are valid inputs:
- An integer e.g.
5. - A list or array of integers
[4, 3, 0]. - A slice object with ints
1:7. - A boolean array.
- A
callable, see Selection By Callable.
In [56]: s1 = pd.Series(np.random.randn(5), index=list(range(0, 10, 2)))In [57]: s1Out[57]:0 0.6957752 0.3417344 0.9597266 -1.1103368 -0.619976dtype: float64In [58]: s1.iloc[:3]Out[58]:0 0.6957752 0.3417344 0.959726dtype: float64In [59]: s1.iloc[3]Out[59]: -1.110336102891167
Note that setting works as well:
In [60]: s1.iloc[:3] = 0In [61]: s1Out[61]:0 0.0000002 0.0000004 0.0000006 -1.1103368 -0.619976dtype: float64
With a DataFrame:
In [62]: df1 = pd.DataFrame(np.random.randn(6, 4),....: index=list(range(0, 12, 2)),....: columns=list(range(0, 8, 2)))....:In [63]: df1Out[63]:0 2 4 60 0.149748 -0.732339 0.687738 0.1764442 0.403310 -0.154951 0.301624 -2.1798614 -1.369849 -0.954208 1.462696 -1.7431616 -0.826591 -0.345352 1.314232 0.6905798 0.995761 2.396780 0.014871 3.35742710 -0.317441 -1.236269 0.896171 -0.487602
Select via integer slicing:
In [64]: df1.iloc[:3]Out[64]:0 2 4 60 0.149748 -0.732339 0.687738 0.1764442 0.403310 -0.154951 0.301624 -2.1798614 -1.369849 -0.954208 1.462696 -1.743161In [65]: df1.iloc[1:5, 2:4]Out[65]:4 62 0.301624 -2.1798614 1.462696 -1.7431616 1.314232 0.6905798 0.014871 3.357427
Select via integer list:
In [66]: df1.iloc[[1, 3, 5], [1, 3]]Out[66]:2 62 -0.154951 -2.1798616 -0.345352 0.69057910 -1.236269 -0.487602
In [67]: df1.iloc[1:3, :]Out[67]:0 2 4 62 0.403310 -0.154951 0.301624 -2.1798614 -1.369849 -0.954208 1.462696 -1.743161
In [68]: df1.iloc[:, 1:3]Out[68]:2 40 -0.732339 0.6877382 -0.154951 0.3016244 -0.954208 1.4626966 -0.345352 1.3142328 2.396780 0.01487110 -1.236269 0.896171
# this is also equivalent to ``df1.iat[1,1]``In [69]: df1.iloc[1, 1]Out[69]: -0.1549507744249032
For getting a cross section using an integer position (equiv to df.xs(1)):
In [70]: df1.iloc[1]Out[70]:0 0.4033102 -0.1549514 0.3016246 -2.179861Name: 2, dtype: float64
Out of range slice indexes are handled gracefully just as in Python/Numpy.
# these are allowed in python/numpy.In [71]: x = list('abcdef')In [72]: xOut[72]: ['a', 'b', 'c', 'd', 'e', 'f']In [73]: x[4:10]Out[73]: ['e', 'f']In [74]: x[8:10]Out[74]: []In [75]: s = pd.Series(x)In [76]: sOut[76]:0 a1 b2 c3 d4 e5 fdtype: objectIn [77]: s.iloc[4:10]Out[77]:4 e5 fdtype: objectIn [78]: s.iloc[8:10]Out[78]: Series([], dtype: object)
Note that using slices that go out of bounds can result in an empty axis (e.g. an empty DataFrame being returned).
In [79]: dfl = pd.DataFrame(np.random.randn(5, 2), columns=list('AB'))In [80]: dflOut[80]:A B0 -0.082240 -2.1829371 0.380396 0.0848442 0.432390 1.5199703 -0.493662 0.6001784 0.274230 0.132885In [81]: dfl.iloc[:, 2:3]Out[81]:Empty DataFrameColumns: []Index: [0, 1, 2, 3, 4]In [82]: dfl.iloc[:, 1:3]Out[82]:B0 -2.1829371 0.0848442 1.5199703 0.6001784 0.132885In [83]: dfl.iloc[4:6]Out[83]:A B4 0.27423 0.132885
A single indexer that is out of bounds will raise an IndexError.
A list of indexers where any element is out of bounds will raise an
IndexError.
>>> dfl.iloc[[4, 5, 6]]IndexError: positional indexers are out-of-bounds>>> dfl.iloc[:, 4]IndexError: single positional indexer is out-of-bounds
Selection by callable
New in version 0.18.1.
.loc, .iloc, and also [] indexing can accept a callable as indexer.
The callable must be a function with one argument (the calling Series or DataFrame) that returns valid output for indexing.
In [84]: df1 = pd.DataFrame(np.random.randn(6, 4),....: index=list('abcdef'),....: columns=list('ABCD'))....:In [85]: df1Out[85]:A B C Da -0.023688 2.410179 1.450520 0.206053b -0.251905 -2.213588 1.063327 1.266143c 0.299368 -0.863838 0.408204 -1.048089d -0.025747 -0.988387 0.094055 1.262731e 1.289997 0.082423 -0.055758 0.536580f -0.489682 0.369374 -0.034571 -2.484478In [86]: df1.loc[lambda df: df.A > 0, :]Out[86]:A B C Dc 0.299368 -0.863838 0.408204 -1.048089e 1.289997 0.082423 -0.055758 0.536580In [87]: df1.loc[:, lambda df: ['A', 'B']]Out[87]:A Ba -0.023688 2.410179b -0.251905 -2.213588c 0.299368 -0.863838d -0.025747 -0.988387e 1.289997 0.082423f -0.489682 0.369374In [88]: df1.iloc[:, lambda df: [0, 1]]Out[88]:A Ba -0.023688 2.410179b -0.251905 -2.213588c 0.299368 -0.863838d -0.025747 -0.988387e 1.289997 0.082423f -0.489682 0.369374In [89]: df1[lambda df: df.columns[0]]Out[89]:a -0.023688b -0.251905c 0.299368d -0.025747e 1.289997f -0.489682Name: A, dtype: float64
You can use callable indexing in Series.
In [90]: df1.A.loc[lambda s: s > 0]Out[90]:c 0.299368e 1.289997Name: A, dtype: float64
Using these methods / indexers, you can chain data selection operations without using a temporary variable.
In [91]: bb = pd.read_csv('data/baseball.csv', index_col='id')In [92]: (bb.groupby(['year', 'team']).sum()....: .loc[lambda df: df.r > 100])....:Out[92]:stint g ab r h X2b X3b hr rbi sb cs bb so ibb hbp sh sf gidpyear team2007 CIN 6 379 745 101 203 35 2 36 125.0 10.0 1.0 105 127.0 14.0 1.0 1.0 15.0 18.0DET 5 301 1062 162 283 54 4 37 144.0 24.0 7.0 97 176.0 3.0 10.0 4.0 8.0 28.0HOU 4 311 926 109 218 47 6 14 77.0 10.0 4.0 60 212.0 3.0 9.0 16.0 6.0 17.0LAN 11 413 1021 153 293 61 3 36 154.0 7.0 5.0 114 141.0 8.0 9.0 3.0 8.0 29.0NYN 13 622 1854 240 509 101 3 61 243.0 22.0 4.0 174 310.0 24.0 23.0 18.0 15.0 48.0SFN 5 482 1305 198 337 67 6 40 171.0 26.0 7.0 235 188.0 51.0 8.0 16.0 6.0 41.0TEX 2 198 729 115 200 40 4 28 115.0 21.0 4.0 73 140.0 4.0 5.0 2.0 8.0 16.0TOR 4 459 1408 187 378 96 2 58 223.0 4.0 2.0 190 265.0 16.0 12.0 4.0 16.0 38.0
IX indexer is deprecated
::: danger Warning
Starting in 0.20.0, the .ix indexer is deprecated, in favor of the more strict .iloc
and .loc indexers.
:::
.ix offers a lot of magic on the inference of what the user wants to do. To wit, .ix can decide
to index positionally OR via labels depending on the data type of the index. This has caused quite a
bit of user confusion over the years.
The recommended methods of indexing are:
.locif you want to label index..ilocif you want to positionally index.
In [93]: dfd = pd.DataFrame({'A': [1, 2, 3],....: 'B': [4, 5, 6]},....: index=list('abc'))....:In [94]: dfdOut[94]:A Ba 1 4b 2 5c 3 6
Previous behavior, where you wish to get the 0th and the 2nd elements from the index in the ‘A’ column.
In [3]: dfd.ix[[0, 2], 'A']Out[3]:a 1c 3Name: A, dtype: int64
Using .loc. Here we will select the appropriate indexes from the index, then use label indexing.
In [95]: dfd.loc[dfd.index[[0, 2]], 'A']Out[95]:a 1c 3Name: A, dtype: int64
This can also be expressed using .iloc, by explicitly getting locations on the indexers, and using
positional indexing to select things.
In [96]: dfd.iloc[[0, 2], dfd.columns.get_loc('A')]Out[96]:a 1c 3Name: A, dtype: int64
For getting multiple indexers, using .get_indexer:
In [97]: dfd.iloc[[0, 2], dfd.columns.get_indexer(['A', 'B'])]Out[97]:A Ba 1 4c 3 6
Indexing with list with missing labels is deprecated
::: danger Warning
Starting in 0.21.0, using .loc or [] with a list with one or more missing labels, is deprecated, in favor of .reindex.
:::
In prior versions, using .loc[list-of-labels] would work as long as at least 1 of the keys was found (otherwise it
would raise a KeyError). This behavior is deprecated and will show a warning message pointing to this section. The
recommended alternative is to use .reindex().
For example.
In [98]: s = pd.Series([1, 2, 3])In [99]: sOut[99]:0 11 22 3dtype: int64
Selection with all keys found is unchanged.
In [100]: s.loc[[1, 2]]Out[100]:1 22 3dtype: int64
Previous behavior
In [4]: s.loc[[1, 2, 3]]Out[4]:1 2.02 3.03 NaNdtype: float64
Current behavior
In [4]: s.loc[[1, 2, 3]]Passing list-likes to .loc with any non-matching elements will raiseKeyError in the future, you can use .reindex() as an alternative.See the documentation here:http://pandas.pydata.org/pandas-docs/stable/indexing.html#deprecate-loc-reindex-listlikeOut[4]:1 2.02 3.03 NaNdtype: float64
Reindexing
The idiomatic way to achieve selecting potentially not-found elements is via .reindex(). See also the section on reindexing.
In [101]: s.reindex([1, 2, 3])Out[101]:1 2.02 3.03 NaNdtype: float64
Alternatively, if you want to select only valid keys, the following is idiomatic and efficient; it is guaranteed to preserve the dtype of the selection.
In [102]: labels = [1, 2, 3]In [103]: s.loc[s.index.intersection(labels)]Out[103]:1 22 3dtype: int64
Having a duplicated index will raise for a .reindex():
In [104]: s = pd.Series(np.arange(4), index=['a', 'a', 'b', 'c'])In [105]: labels = ['c', 'd']
In [17]: s.reindex(labels)ValueError: cannot reindex from a duplicate axis
Generally, you can intersect the desired labels with the current axis, and then reindex.
In [106]: s.loc[s.index.intersection(labels)].reindex(labels)Out[106]:c 3.0d NaNdtype: float64
However, this would still raise if your resulting index is duplicated.
In [41]: labels = ['a', 'd']In [42]: s.loc[s.index.intersection(labels)].reindex(labels)ValueError: cannot reindex from a duplicate axis
Selecting random samples
A random selection of rows or columns from a Series or DataFrame with the sample() method. The method will sample rows by default, and accepts a specific number of rows/columns to return, or a fraction of rows.
In [107]: s = pd.Series([0, 1, 2, 3, 4, 5])# When no arguments are passed, returns 1 row.In [108]: s.sample()Out[108]:4 4dtype: int64# One may specify either a number of rows:In [109]: s.sample(n=3)Out[109]:0 04 41 1dtype: int64# Or a fraction of the rows:In [110]: s.sample(frac=0.5)Out[110]:5 53 31 1dtype: int64
By default, sample will return each row at most once, but one can also sample with replacement
using the replace option:
In [111]: s = pd.Series([0, 1, 2, 3, 4, 5])# Without replacement (default):In [112]: s.sample(n=6, replace=False)Out[112]:0 01 15 53 32 24 4dtype: int64# With replacement:In [113]: s.sample(n=6, replace=True)Out[113]:0 04 43 32 24 44 4dtype: int64
By default, each row has an equal probability of being selected, but if you want rows
to have different probabilities, you can pass the sample function sampling weights as
weights. These weights can be a list, a NumPy array, or a Series, but they must be of the same length as the object you are sampling. Missing values will be treated as a weight of zero, and inf values are not allowed. If weights do not sum to 1, they will be re-normalized by dividing all weights by the sum of the weights. For example:
In [114]: s = pd.Series([0, 1, 2, 3, 4, 5])In [115]: example_weights = [0, 0, 0.2, 0.2, 0.2, 0.4]In [116]: s.sample(n=3, weights=example_weights)Out[116]:5 54 43 3dtype: int64# Weights will be re-normalized automaticallyIn [117]: example_weights2 = [0.5, 0, 0, 0, 0, 0]In [118]: s.sample(n=1, weights=example_weights2)Out[118]:0 0dtype: int64
When applied to a DataFrame, you can use a column of the DataFrame as sampling weights (provided you are sampling rows and not columns) by simply passing the name of the column as a string.
In [119]: df2 = pd.DataFrame({'col1': [9, 8, 7, 6],.....: 'weight_column': [0.5, 0.4, 0.1, 0]}).....:In [120]: df2.sample(n=3, weights='weight_column')Out[120]:col1 weight_column1 8 0.40 9 0.52 7 0.1
sample also allows users to sample columns instead of rows using the axis argument.
In [121]: df3 = pd.DataFrame({'col1': [1, 2, 3], 'col2': [2, 3, 4]})In [122]: df3.sample(n=1, axis=1)Out[122]:col10 11 22 3
Finally, one can also set a seed for sample’s random number generator using the random_state argument, which will accept either an integer (as a seed) or a NumPy RandomState object.
In [123]: df4 = pd.DataFrame({'col1': [1, 2, 3], 'col2': [2, 3, 4]})# With a given seed, the sample will always draw the same rows.In [124]: df4.sample(n=2, random_state=2)Out[124]:col1 col22 3 41 2 3In [125]: df4.sample(n=2, random_state=2)Out[125]:col1 col22 3 41 2 3
Setting with enlargement
The .loc/[] operations can perform enlargement when setting a non-existent key for that axis.
In the Series case this is effectively an appending operation.
In [126]: se = pd.Series([1, 2, 3])In [127]: seOut[127]:0 11 22 3dtype: int64In [128]: se[5] = 5.In [129]: seOut[129]:0 1.01 2.02 3.05 5.0dtype: float64
A DataFrame can be enlarged on either axis via .loc.
In [130]: dfi = pd.DataFrame(np.arange(6).reshape(3, 2),.....: columns=['A', 'B']).....:In [131]: dfiOut[131]:A B0 0 11 2 32 4 5In [132]: dfi.loc[:, 'C'] = dfi.loc[:, 'A']In [133]: dfiOut[133]:A B C0 0 1 01 2 3 22 4 5 4
This is like an append operation on the DataFrame.
In [134]: dfi.loc[3] = 5In [135]: dfiOut[135]:A B C0 0 1 01 2 3 22 4 5 43 5 5 5
Fast scalar value getting and setting
Since indexing with [] must handle a lot of cases (single-label access,
slicing, boolean indexing, etc.), it has a bit of overhead in order to figure
out what you’re asking for. If you only want to access a scalar value, the
fastest way is to use the at and iat methods, which are implemented on
all of the data structures.
Similarly to loc, at provides label based scalar lookups, while, iat provides integer based lookups analogously to iloc
In [136]: s.iat[5]Out[136]: 5In [137]: df.at[dates[5], 'A']Out[137]: -0.6736897080883706In [138]: df.iat[3, 0]Out[138]: 0.7215551622443669
You can also set using these same indexers.
In [139]: df.at[dates[5], 'E'] = 7In [140]: df.iat[3, 0] = 7
at may enlarge the object in-place as above if the indexer is missing.
In [141]: df.at[dates[-1] + pd.Timedelta('1 day'), 0] = 7In [142]: dfOut[142]:A B C D E 02000-01-01 0.469112 -0.282863 -1.509059 -1.135632 NaN NaN2000-01-02 1.212112 -0.173215 0.119209 -1.044236 NaN NaN2000-01-03 -0.861849 -2.104569 -0.494929 1.071804 NaN NaN2000-01-04 7.000000 -0.706771 -1.039575 0.271860 NaN NaN2000-01-05 -0.424972 0.567020 0.276232 -1.087401 NaN NaN2000-01-06 -0.673690 0.113648 -1.478427 0.524988 7.0 NaN2000-01-07 0.404705 0.577046 -1.715002 -1.039268 NaN NaN2000-01-08 -0.370647 -1.157892 -1.344312 0.844885 NaN NaN2000-01-09 NaN NaN NaN NaN NaN 7.0
Boolean indexing
Another common operation is the use of boolean vectors to filter the data.
The operators are: | for or, & for and, and ~ for not.
These must be grouped by using parentheses, since by default Python will
evaluate an expression such as df.A > 2 & df.B < 3 as
df.A > (2 & df.B) < 3, while the desired evaluation order is
(df.A > 2) & (df.B < 3).
Using a boolean vector to index a Series works exactly as in a NumPy ndarray:
In [143]: s = pd.Series(range(-3, 4))In [144]: sOut[144]:0 -31 -22 -13 04 15 26 3dtype: int64In [145]: s[s > 0]Out[145]:4 15 26 3dtype: int64In [146]: s[(s < -1) | (s > 0.5)]Out[146]:0 -31 -24 15 26 3dtype: int64In [147]: s[~(s < 0)]Out[147]:3 04 15 26 3dtype: int64
You may select rows from a DataFrame using a boolean vector the same length as the DataFrame’s index (for example, something derived from one of the columns of the DataFrame):
In [148]: df[df['A'] > 0]Out[148]:A B C D E 02000-01-01 0.469112 -0.282863 -1.509059 -1.135632 NaN NaN2000-01-02 1.212112 -0.173215 0.119209 -1.044236 NaN NaN2000-01-04 7.000000 -0.706771 -1.039575 0.271860 NaN NaN2000-01-07 0.404705 0.577046 -1.715002 -1.039268 NaN NaN
List comprehensions and the map method of Series can also be used to produce
more complex criteria:
In [149]: df2 = pd.DataFrame({'a': ['one', 'one', 'two', 'three', 'two', 'one', 'six'],.....: 'b': ['x', 'y', 'y', 'x', 'y', 'x', 'x'],.....: 'c': np.random.randn(7)}).....:# only want 'two' or 'three'In [150]: criterion = df2['a'].map(lambda x: x.startswith('t'))In [151]: df2[criterion]Out[151]:a b c2 two y 0.0412903 three x 0.3617194 two y -0.238075# equivalent but slowerIn [152]: df2[[x.startswith('t') for x in df2['a']]]Out[152]:a b c2 two y 0.0412903 three x 0.3617194 two y -0.238075# Multiple criteriaIn [153]: df2[criterion & (df2['b'] == 'x')]Out[153]:a b c3 three x 0.361719
With the choice methods Selection by Label, Selection by Position, and Advanced Indexing you may select along more than one axis using boolean vectors combined with other indexing expressions.
In [154]: df2.loc[criterion & (df2['b'] == 'x'), 'b':'c']Out[154]:b c3 x 0.361719
Indexing with isin
Consider the isin() method of Series, which returns a boolean
vector that is true wherever the Series elements exist in the passed list.
This allows you to select rows where one or more columns have values you want:
In [155]: s = pd.Series(np.arange(5), index=np.arange(5)[::-1], dtype='int64')In [156]: sOut[156]:4 03 12 21 30 4dtype: int64In [157]: s.isin([2, 4, 6])Out[157]:4 False3 False2 True1 False0 Truedtype: boolIn [158]: s[s.isin([2, 4, 6])]Out[158]:2 20 4dtype: int64
The same method is available for Index objects and is useful for the cases
when you don’t know which of the sought labels are in fact present:
In [159]: s[s.index.isin([2, 4, 6])]Out[159]:4 02 2dtype: int64# compare it to the followingIn [160]: s.reindex([2, 4, 6])Out[160]:2 2.04 0.06 NaNdtype: float64
In addition to that, MultiIndex allows selecting a separate level to use
in the membership check:
In [161]: s_mi = pd.Series(np.arange(6),.....: index=pd.MultiIndex.from_product([[0, 1], ['a', 'b', 'c']])).....:In [162]: s_miOut[162]:0 a 0b 1c 21 a 3b 4c 5dtype: int64In [163]: s_mi.iloc[s_mi.index.isin([(1, 'a'), (2, 'b'), (0, 'c')])]Out[163]:0 c 21 a 3dtype: int64In [164]: s_mi.iloc[s_mi.index.isin(['a', 'c', 'e'], level=1)]Out[164]:0 a 0c 21 a 3c 5dtype: int64
DataFrame also has an isin() method. When calling isin, pass a set of
values as either an array or dict. If values is an array, isin returns
a DataFrame of booleans that is the same shape as the original DataFrame, with True
wherever the element is in the sequence of values.
In [165]: df = pd.DataFrame({'vals': [1, 2, 3, 4], 'ids': ['a', 'b', 'f', 'n'],.....: 'ids2': ['a', 'n', 'c', 'n']}).....:In [166]: values = ['a', 'b', 1, 3]In [167]: df.isin(values)Out[167]:vals ids ids20 True True True1 False True False2 True False False3 False False False
Oftentimes you’ll want to match certain values with certain columns.
Just make values a dict where the key is the column, and the value is
a list of items you want to check for.
In [168]: values = {'ids': ['a', 'b'], 'vals': [1, 3]}In [169]: df.isin(values)Out[169]:vals ids ids20 True True False1 False True False2 True False False3 False False False
Combine DataFrame’s isin with the any() and all() methods to
quickly select subsets of your data that meet a given criteria.
To select a row where each column meets its own criterion:
In [170]: values = {'ids': ['a', 'b'], 'ids2': ['a', 'c'], 'vals': [1, 3]}In [171]: row_mask = df.isin(values).all(1)In [172]: df[row_mask]Out[172]:vals ids ids20 1 a a
The where() Method and Masking
Selecting values from a Series with a boolean vector generally returns a
subset of the data. To guarantee that selection output has the same shape as
the original data, you can use the where method in Series and DataFrame.
To return only the selected rows:
In [173]: s[s > 0]Out[173]:3 12 21 30 4dtype: int64
To return a Series of the same shape as the original:
In [174]: s.where(s > 0)Out[174]:4 NaN3 1.02 2.01 3.00 4.0dtype: float64
Selecting values from a DataFrame with a boolean criterion now also preserves
input data shape. where is used under the hood as the implementation.
The code below is equivalent to df.where(df < 0).
In [175]: df[df < 0]Out[175]:A B C D2000-01-01 -2.104139 -1.309525 NaN NaN2000-01-02 -0.352480 NaN -1.192319 NaN2000-01-03 -0.864883 NaN -0.227870 NaN2000-01-04 NaN -1.222082 NaN -1.2332032000-01-05 NaN -0.605656 -1.169184 NaN2000-01-06 NaN -0.948458 NaN -0.6847182000-01-07 -2.670153 -0.114722 NaN -0.0480482000-01-08 NaN NaN -0.048788 -0.808838
In addition, where takes an optional other argument for replacement of
values where the condition is False, in the returned copy.
In [176]: df.where(df < 0, -df)Out[176]:A B C D2000-01-01 -2.104139 -1.309525 -0.485855 -0.2451662000-01-02 -0.352480 -0.390389 -1.192319 -1.6558242000-01-03 -0.864883 -0.299674 -0.227870 -0.2810592000-01-04 -0.846958 -1.222082 -0.600705 -1.2332032000-01-05 -0.669692 -0.605656 -1.169184 -0.3424162000-01-06 -0.868584 -0.948458 -2.297780 -0.6847182000-01-07 -2.670153 -0.114722 -0.168904 -0.0480482000-01-08 -0.801196 -1.392071 -0.048788 -0.808838
You may wish to set values based on some boolean criteria. This can be done intuitively like so:
In [177]: s2 = s.copy()In [178]: s2[s2 < 0] = 0In [179]: s2Out[179]:4 03 12 21 30 4dtype: int64In [180]: df2 = df.copy()In [181]: df2[df2 < 0] = 0In [182]: df2Out[182]:A B C D2000-01-01 0.000000 0.000000 0.485855 0.2451662000-01-02 0.000000 0.390389 0.000000 1.6558242000-01-03 0.000000 0.299674 0.000000 0.2810592000-01-04 0.846958 0.000000 0.600705 0.0000002000-01-05 0.669692 0.000000 0.000000 0.3424162000-01-06 0.868584 0.000000 2.297780 0.0000002000-01-07 0.000000 0.000000 0.168904 0.0000002000-01-08 0.801196 1.392071 0.000000 0.000000
By default, where returns a modified copy of the data. There is an
optional parameter inplace so that the original data can be modified
without creating a copy:
In [183]: df_orig = df.copy()In [184]: df_orig.where(df > 0, -df, inplace=True)In [185]: df_origOut[185]:A B C D2000-01-01 2.104139 1.309525 0.485855 0.2451662000-01-02 0.352480 0.390389 1.192319 1.6558242000-01-03 0.864883 0.299674 0.227870 0.2810592000-01-04 0.846958 1.222082 0.600705 1.2332032000-01-05 0.669692 0.605656 1.169184 0.3424162000-01-06 0.868584 0.948458 2.297780 0.6847182000-01-07 2.670153 0.114722 0.168904 0.0480482000-01-08 0.801196 1.392071 0.048788 0.808838
::: tip Note
The signature for DataFrame.where() differs from numpy.where().
Roughly df1.where(m, df2) is equivalent to np.where(m, df1, df2).
In [186]: df.where(df < 0, -df) == np.where(df < 0, df, -df)Out[186]:A B C D2000-01-01 True True True True2000-01-02 True True True True2000-01-03 True True True True2000-01-04 True True True True2000-01-05 True True True True2000-01-06 True True True True2000-01-07 True True True True2000-01-08 True True True True
:::
Alignment
Furthermore, where aligns the input boolean condition (ndarray or DataFrame),
such that partial selection with setting is possible. This is analogous to
partial setting via .loc (but on the contents rather than the axis labels).
In [187]: df2 = df.copy()In [188]: df2[df2[1:4] > 0] = 3In [189]: df2Out[189]:A B C D2000-01-01 -2.104139 -1.309525 0.485855 0.2451662000-01-02 -0.352480 3.000000 -1.192319 3.0000002000-01-03 -0.864883 3.000000 -0.227870 3.0000002000-01-04 3.000000 -1.222082 3.000000 -1.2332032000-01-05 0.669692 -0.605656 -1.169184 0.3424162000-01-06 0.868584 -0.948458 2.297780 -0.6847182000-01-07 -2.670153 -0.114722 0.168904 -0.0480482000-01-08 0.801196 1.392071 -0.048788 -0.808838
Where can also accept axis and level parameters to align the input when
performing the where.
In [190]: df2 = df.copy()In [191]: df2.where(df2 > 0, df2['A'], axis='index')Out[191]:A B C D2000-01-01 -2.104139 -2.104139 0.485855 0.2451662000-01-02 -0.352480 0.390389 -0.352480 1.6558242000-01-03 -0.864883 0.299674 -0.864883 0.2810592000-01-04 0.846958 0.846958 0.600705 0.8469582000-01-05 0.669692 0.669692 0.669692 0.3424162000-01-06 0.868584 0.868584 2.297780 0.8685842000-01-07 -2.670153 -2.670153 0.168904 -2.6701532000-01-08 0.801196 1.392071 0.801196 0.801196
This is equivalent to (but faster than) the following.
In [192]: df2 = df.copy()In [193]: df.apply(lambda x, y: x.where(x > 0, y), y=df['A'])Out[193]:A B C D2000-01-01 -2.104139 -2.104139 0.485855 0.2451662000-01-02 -0.352480 0.390389 -0.352480 1.6558242000-01-03 -0.864883 0.299674 -0.864883 0.2810592000-01-04 0.846958 0.846958 0.600705 0.8469582000-01-05 0.669692 0.669692 0.669692 0.3424162000-01-06 0.868584 0.868584 2.297780 0.8685842000-01-07 -2.670153 -2.670153 0.168904 -2.6701532000-01-08 0.801196 1.392071 0.801196 0.801196
New in version 0.18.1.
Where can accept a callable as condition and other arguments. The function must
be with one argument (the calling Series or DataFrame) and that returns valid output
as condition and other argument.
In [194]: df3 = pd.DataFrame({'A': [1, 2, 3],.....: 'B': [4, 5, 6],.....: 'C': [7, 8, 9]}).....:In [195]: df3.where(lambda x: x > 4, lambda x: x + 10)Out[195]:A B C0 11 14 71 12 5 82 13 6 9
Mask
mask() is the inverse boolean operation of where.
In [196]: s.mask(s >= 0)Out[196]:4 NaN3 NaN2 NaN1 NaN0 NaNdtype: float64In [197]: df.mask(df >= 0)Out[197]:A B C D2000-01-01 -2.104139 -1.309525 NaN NaN2000-01-02 -0.352480 NaN -1.192319 NaN2000-01-03 -0.864883 NaN -0.227870 NaN2000-01-04 NaN -1.222082 NaN -1.2332032000-01-05 NaN -0.605656 -1.169184 NaN2000-01-06 NaN -0.948458 NaN -0.6847182000-01-07 -2.670153 -0.114722 NaN -0.0480482000-01-08 NaN NaN -0.048788 -0.808838
The query() Method
DataFrame objects have a query()
method that allows selection using an expression.
You can get the value of the frame where column b has values
between the values of columns a and c. For example:
In [198]: n = 10In [199]: df = pd.DataFrame(np.random.rand(n, 3), columns=list('abc'))In [200]: dfOut[200]:a b c0 0.438921 0.118680 0.8636701 0.138138 0.577363 0.6866022 0.595307 0.564592 0.5206303 0.913052 0.926075 0.6161844 0.078718 0.854477 0.8987255 0.076404 0.523211 0.5915386 0.792342 0.216974 0.5640567 0.397890 0.454131 0.9157168 0.074315 0.437913 0.0197949 0.559209 0.502065 0.026437# pure pythonIn [201]: df[(df.a < df.b) & (df.b < df.c)]Out[201]:a b c1 0.138138 0.577363 0.6866024 0.078718 0.854477 0.8987255 0.076404 0.523211 0.5915387 0.397890 0.454131 0.915716# queryIn [202]: df.query('(a < b) & (b < c)')Out[202]:a b c1 0.138138 0.577363 0.6866024 0.078718 0.854477 0.8987255 0.076404 0.523211 0.5915387 0.397890 0.454131 0.915716
Do the same thing but fall back on a named index if there is no column
with the name a.
In [203]: df = pd.DataFrame(np.random.randint(n / 2, size=(n, 2)), columns=list('bc'))In [204]: df.index.name = 'a'In [205]: dfOut[205]:b ca0 0 41 0 12 3 43 4 34 1 45 0 36 0 17 3 48 2 39 1 1In [206]: df.query('a < b and b < c')Out[206]:b ca2 3 4
If instead you don’t want to or cannot name your index, you can use the name
index in your query expression:
In [207]: df = pd.DataFrame(np.random.randint(n, size=(n, 2)), columns=list('bc'))In [208]: dfOut[208]:b c0 3 11 3 02 5 63 5 24 7 45 0 16 2 57 0 18 6 09 7 9In [209]: df.query('index < b < c')Out[209]:b c2 5 6
::: tip Note
If the name of your index overlaps with a column name, the column name is given precedence. For example,
In [210]: df = pd.DataFrame({'a': np.random.randint(5, size=5)})In [211]: df.index.name = 'a'In [212]: df.query('a > 2') # uses the column 'a', not the indexOut[212]:aa1 33 3
You can still use the index in a query expression by using the special identifier ‘index’:
In [213]: df.query('index > 2')Out[213]:aa3 34 2
If for some reason you have a column named index, then you can refer to
the index as ilevel_0 as well, but at this point you should consider
renaming your columns to something less ambiguous.
:::
MultiIndex query() Syntax
You can also use the levels of a DataFrame with a
MultiIndex as if they were columns in the frame:
In [214]: n = 10In [215]: colors = np.random.choice(['red', 'green'], size=n)In [216]: foods = np.random.choice(['eggs', 'ham'], size=n)In [217]: colorsOut[217]:array(['red', 'red', 'red', 'green', 'green', 'green', 'green', 'green','green', 'green'], dtype='<U5')In [218]: foodsOut[218]:array(['ham', 'ham', 'eggs', 'eggs', 'eggs', 'ham', 'ham', 'eggs', 'eggs','eggs'], dtype='<U4')In [219]: index = pd.MultiIndex.from_arrays([colors, foods], names=['color', 'food'])In [220]: df = pd.DataFrame(np.random.randn(n, 2), index=index)In [221]: dfOut[221]:0 1color foodred ham 0.194889 -0.381994ham 0.318587 2.089075eggs -0.728293 -0.090255green eggs -0.748199 1.318931eggs -2.029766 0.792652ham 0.461007 -0.542749ham -0.305384 -0.479195eggs 0.095031 -0.270099eggs -0.707140 -0.773882eggs 0.229453 0.304418In [222]: df.query('color == "red"')Out[222]:0 1color foodred ham 0.194889 -0.381994ham 0.318587 2.089075eggs -0.728293 -0.090255
If the levels of the MultiIndex are unnamed, you can refer to them using
special names:
In [223]: df.index.names = [None, None]In [224]: dfOut[224]:0 1red ham 0.194889 -0.381994ham 0.318587 2.089075eggs -0.728293 -0.090255green eggs -0.748199 1.318931eggs -2.029766 0.792652ham 0.461007 -0.542749ham -0.305384 -0.479195eggs 0.095031 -0.270099eggs -0.707140 -0.773882eggs 0.229453 0.304418In [225]: df.query('ilevel_0 == "red"')Out[225]:0 1red ham 0.194889 -0.381994ham 0.318587 2.089075eggs -0.728293 -0.090255
The convention is ilevel_0, which means “index level 0” for the 0th level
of the index.
query() Use Cases
A use case for query() is when you have a collection of
DataFrame objects that have a subset of column names (or index
levels/names) in common. You can pass the same query to both frames without
having to specify which frame you’re interested in querying
In [226]: df = pd.DataFrame(np.random.rand(n, 3), columns=list('abc'))In [227]: dfOut[227]:a b c0 0.224283 0.736107 0.1391681 0.302827 0.657803 0.7138972 0.611185 0.136624 0.9849603 0.195246 0.123436 0.6277124 0.618673 0.371660 0.0479025 0.480088 0.062993 0.1857606 0.568018 0.483467 0.4452897 0.309040 0.274580 0.5871018 0.258993 0.477769 0.3702559 0.550459 0.840870 0.304611In [228]: df2 = pd.DataFrame(np.random.rand(n + 2, 3), columns=df.columns)In [229]: df2Out[229]:a b c0 0.357579 0.229800 0.5960011 0.309059 0.957923 0.9656632 0.123102 0.336914 0.3186163 0.526506 0.323321 0.8608134 0.518736 0.486514 0.3847245 0.190804 0.505723 0.6145336 0.891939 0.623977 0.6766397 0.480559 0.378528 0.4608588 0.420223 0.136404 0.1412959 0.732206 0.419540 0.60467510 0.604466 0.848974 0.89616511 0.589168 0.920046 0.732716In [230]: expr = '0.0 <= a <= c <= 0.5'In [231]: map(lambda frame: frame.query(expr), [df, df2])Out[231]: <map at 0x7f65f7952d30>
query() Python versus pandas Syntax Comparison
Full numpy-like syntax:
In [232]: df = pd.DataFrame(np.random.randint(n, size=(n, 3)), columns=list('abc'))In [233]: dfOut[233]:a b c0 7 8 91 1 0 72 2 7 23 6 2 24 2 6 35 3 8 26 1 7 27 5 1 58 9 8 09 1 5 0In [234]: df.query('(a < b) & (b < c)')Out[234]:a b c0 7 8 9In [235]: df[(df.a < df.b) & (df.b < df.c)]Out[235]:a b c0 7 8 9
Slightly nicer by removing the parentheses (by binding making comparison
operators bind tighter than & and |).
In [236]: df.query('a < b & b < c')Out[236]:a b c0 7 8 9
Use English instead of symbols:
In [237]: df.query('a < b and b < c')Out[237]:a b c0 7 8 9
Pretty close to how you might write it on paper:
In [238]: df.query('a < b < c')Out[238]:a b c0 7 8 9
The in and not in operators
query() also supports special use of Python’s in and
not in comparison operators, providing a succinct syntax for calling the
isin method of a Series or DataFrame.
# get all rows where columns "a" and "b" have overlapping valuesIn [239]: df = pd.DataFrame({'a': list('aabbccddeeff'), 'b': list('aaaabbbbcccc'),.....: 'c': np.random.randint(5, size=12),.....: 'd': np.random.randint(9, size=12)}).....:In [240]: dfOut[240]:a b c d0 a a 2 61 a a 4 72 b a 1 63 b a 2 14 c b 3 65 c b 0 26 d b 3 37 d b 2 18 e c 4 39 e c 2 010 f c 0 611 f c 1 2In [241]: df.query('a in b')Out[241]:a b c d0 a a 2 61 a a 4 72 b a 1 63 b a 2 14 c b 3 65 c b 0 2# How you'd do it in pure PythonIn [242]: df[df.a.isin(df.b)]Out[242]:a b c d0 a a 2 61 a a 4 72 b a 1 63 b a 2 14 c b 3 65 c b 0 2In [243]: df.query('a not in b')Out[243]:a b c d6 d b 3 37 d b 2 18 e c 4 39 e c 2 010 f c 0 611 f c 1 2# pure PythonIn [244]: df[~df.a.isin(df.b)]Out[244]:a b c d6 d b 3 37 d b 2 18 e c 4 39 e c 2 010 f c 0 611 f c 1 2
You can combine this with other expressions for very succinct queries:
# rows where cols a and b have overlapping values# and col c's values are less than col d'sIn [245]: df.query('a in b and c < d')Out[245]:a b c d0 a a 2 61 a a 4 72 b a 1 64 c b 3 65 c b 0 2# pure PythonIn [246]: df[df.b.isin(df.a) & (df.c < df.d)]Out[246]:a b c d0 a a 2 61 a a 4 72 b a 1 64 c b 3 65 c b 0 210 f c 0 611 f c 1 2
::: tip Note
Note that in and not in are evaluated in Python, since numexpr
has no equivalent of this operation. However, only the in/not in
expression itself is evaluated in vanilla Python. For example, in the
expression
df.query('a in b + c + d')
(b + c + d) is evaluated by numexpr and then the in
operation is evaluated in plain Python. In general, any operations that can
be evaluated using numexpr will be.
:::
Special use of the == operator with list objects
Comparing a list of values to a column using ==/!= works similarly
to in/not in.
In [247]: df.query('b == ["a", "b", "c"]')Out[247]:a b c d0 a a 2 61 a a 4 72 b a 1 63 b a 2 14 c b 3 65 c b 0 26 d b 3 37 d b 2 18 e c 4 39 e c 2 010 f c 0 611 f c 1 2# pure PythonIn [248]: df[df.b.isin(["a", "b", "c"])]Out[248]:a b c d0 a a 2 61 a a 4 72 b a 1 63 b a 2 14 c b 3 65 c b 0 26 d b 3 37 d b 2 18 e c 4 39 e c 2 010 f c 0 611 f c 1 2In [249]: df.query('c == [1, 2]')Out[249]:a b c d0 a a 2 62 b a 1 63 b a 2 17 d b 2 19 e c 2 011 f c 1 2In [250]: df.query('c != [1, 2]')Out[250]:a b c d1 a a 4 74 c b 3 65 c b 0 26 d b 3 38 e c 4 310 f c 0 6# using in/not inIn [251]: df.query('[1, 2] in c')Out[251]:a b c d0 a a 2 62 b a 1 63 b a 2 17 d b 2 19 e c 2 011 f c 1 2In [252]: df.query('[1, 2] not in c')Out[252]:a b c d1 a a 4 74 c b 3 65 c b 0 26 d b 3 38 e c 4 310 f c 0 6# pure PythonIn [253]: df[df.c.isin([1, 2])]Out[253]:a b c d0 a a 2 62 b a 1 63 b a 2 17 d b 2 19 e c 2 011 f c 1 2
Boolean operators
You can negate boolean expressions with the word not or the ~ operator.
In [254]: df = pd.DataFrame(np.random.rand(n, 3), columns=list('abc'))In [255]: df['bools'] = np.random.rand(len(df)) > 0.5In [256]: df.query('~bools')Out[256]:a b c bools2 0.697753 0.212799 0.329209 False7 0.275396 0.691034 0.826619 False8 0.190649 0.558748 0.262467 FalseIn [257]: df.query('not bools')Out[257]:a b c bools2 0.697753 0.212799 0.329209 False7 0.275396 0.691034 0.826619 False8 0.190649 0.558748 0.262467 FalseIn [258]: df.query('not bools') == df[~df.bools]Out[258]:a b c bools2 True True True True7 True True True True8 True True True True
Of course, expressions can be arbitrarily complex too:
# short query syntaxIn [259]: shorter = df.query('a < b < c and (not bools) or bools > 2')# equivalent in pure PythonIn [260]: longer = df[(df.a < df.b) & (df.b < df.c) & (~df.bools) | (df.bools > 2)]In [261]: shorterOut[261]:a b c bools7 0.275396 0.691034 0.826619 FalseIn [262]: longerOut[262]:a b c bools7 0.275396 0.691034 0.826619 FalseIn [263]: shorter == longerOut[263]:a b c bools7 True True True True
Performance of query()
DataFrame.query() using numexpr is slightly faster than Python for
large frames.
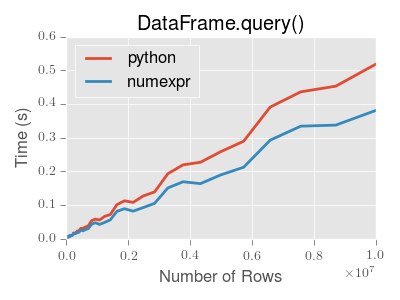
::: tip Note
You will only see the performance benefits of using the numexpr engine
with DataFrame.query() if your frame has more than approximately 200,000
rows.
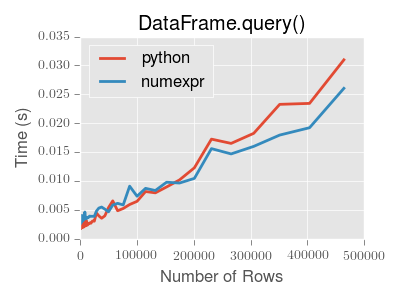
:::
This plot was created using a DataFrame with 3 columns each containing
floating point values generated using numpy.random.randn().
Duplicate data
If you want to identify and remove duplicate rows in a DataFrame, there are
two methods that will help: duplicated and drop_duplicates. Each
takes as an argument the columns to use to identify duplicated rows.
duplicatedreturns a boolean vector whose length is the number of rows, and which indicates whether a row is duplicated.drop_duplicatesremoves duplicate rows.
By default, the first observed row of a duplicate set is considered unique, but
each method has a keep parameter to specify targets to be kept.
keep='first'(default): mark / drop duplicates except for the first occurrence.keep='last': mark / drop duplicates except for the last occurrence.keep=False: mark / drop all duplicates.
In [264]: df2 = pd.DataFrame({'a': ['one', 'one', 'two', 'two', 'two', 'three', 'four'],.....: 'b': ['x', 'y', 'x', 'y', 'x', 'x', 'x'],.....: 'c': np.random.randn(7)}).....:In [265]: df2Out[265]:a b c0 one x -1.0671371 one y 0.3095002 two x -0.2110563 two y -1.8420234 two x -0.3908205 three x -1.9644756 four x 1.298329In [266]: df2.duplicated('a')Out[266]:0 False1 True2 False3 True4 True5 False6 Falsedtype: boolIn [267]: df2.duplicated('a', keep='last')Out[267]:0 True1 False2 True3 True4 False5 False6 Falsedtype: boolIn [268]: df2.duplicated('a', keep=False)Out[268]:0 True1 True2 True3 True4 True5 False6 Falsedtype: boolIn [269]: df2.drop_duplicates('a')Out[269]:a b c0 one x -1.0671372 two x -0.2110565 three x -1.9644756 four x 1.298329In [270]: df2.drop_duplicates('a', keep='last')Out[270]:a b c1 one y 0.3095004 two x -0.3908205 three x -1.9644756 four x 1.298329In [271]: df2.drop_duplicates('a', keep=False)Out[271]:a b c5 three x -1.9644756 four x 1.298329
Also, you can pass a list of columns to identify duplications.
In [272]: df2.duplicated(['a', 'b'])Out[272]:0 False1 False2 False3 False4 True5 False6 Falsedtype: boolIn [273]: df2.drop_duplicates(['a', 'b'])Out[273]:a b c0 one x -1.0671371 one y 0.3095002 two x -0.2110563 two y -1.8420235 three x -1.9644756 four x 1.298329
To drop duplicates by index value, use Index.duplicated then perform slicing.
The same set of options are available for the keep parameter.
In [274]: df3 = pd.DataFrame({'a': np.arange(6),.....: 'b': np.random.randn(6)},.....: index=['a', 'a', 'b', 'c', 'b', 'a']).....:In [275]: df3Out[275]:a ba 0 1.440455a 1 2.456086b 2 1.038402c 3 -0.894409b 4 0.683536a 5 3.082764In [276]: df3.index.duplicated()Out[276]: array([False, True, False, False, True, True])In [277]: df3[~df3.index.duplicated()]Out[277]:a ba 0 1.440455b 2 1.038402c 3 -0.894409In [278]: df3[~df3.index.duplicated(keep='last')]Out[278]:a bc 3 -0.894409b 4 0.683536a 5 3.082764In [279]: df3[~df3.index.duplicated(keep=False)]Out[279]:a bc 3 -0.894409
Dictionary-like get() method
Each of Series or DataFrame have a get method which can return a
default value.
In [280]: s = pd.Series([1, 2, 3], index=['a', 'b', 'c'])In [281]: s.get('a') # equivalent to s['a']Out[281]: 1In [282]: s.get('x', default=-1)Out[282]: -1
The lookup() method
Sometimes you want to extract a set of values given a sequence of row labels
and column labels, and the lookup method allows for this and returns a
NumPy array. For instance:
In [283]: dflookup = pd.DataFrame(np.random.rand(20, 4), columns = ['A', 'B', 'C', 'D'])In [284]: dflookup.lookup(list(range(0, 10, 2)), ['B', 'C', 'A', 'B', 'D'])Out[284]: array([0.3506, 0.4779, 0.4825, 0.9197, 0.5019])
Index objects
The pandas Index class and its subclasses can be viewed as
implementing an ordered multiset. Duplicates are allowed. However, if you try
to convert an Index object with duplicate entries into a
set, an exception will be raised.
Index also provides the infrastructure necessary for
lookups, data alignment, and reindexing. The easiest way to create an
Index directly is to pass a list or other sequence to
Index:
In [285]: index = pd.Index(['e', 'd', 'a', 'b'])In [286]: indexOut[286]: Index(['e', 'd', 'a', 'b'], dtype='object')In [287]: 'd' in indexOut[287]: True
You can also pass a name to be stored in the index:
In [288]: index = pd.Index(['e', 'd', 'a', 'b'], name='something')In [289]: index.nameOut[289]: 'something'
The name, if set, will be shown in the console display:
In [290]: index = pd.Index(list(range(5)), name='rows')In [291]: columns = pd.Index(['A', 'B', 'C'], name='cols')In [292]: df = pd.DataFrame(np.random.randn(5, 3), index=index, columns=columns)In [293]: dfOut[293]:cols A B Crows0 1.295989 0.185778 0.4362591 0.678101 0.311369 -0.5283782 -0.674808 -1.103529 -0.6561573 1.889957 2.076651 -1.1021924 -1.211795 -0.791746 0.634724In [294]: df['A']Out[294]:rows0 1.2959891 0.6781012 -0.6748083 1.8899574 -1.211795Name: A, dtype: float64
Setting metadata
Indexes are “mostly immutable”, but it is possible to set and change their
metadata, like the index name (or, for MultiIndex, levels and
codes).
You can use the rename, set_names, set_levels, and set_codes
to set these attributes directly. They default to returning a copy; however,
you can specify inplace=True to have the data change in place.
See Advanced Indexing for usage of MultiIndexes.
In [295]: ind = pd.Index([1, 2, 3])In [296]: ind.rename("apple")Out[296]: Int64Index([1, 2, 3], dtype='int64', name='apple')In [297]: indOut[297]: Int64Index([1, 2, 3], dtype='int64')In [298]: ind.set_names(["apple"], inplace=True)In [299]: ind.name = "bob"In [300]: indOut[300]: Int64Index([1, 2, 3], dtype='int64', name='bob')
set_names, set_levels, and set_codes also take an optional
level argument
In [301]: index = pd.MultiIndex.from_product([range(3), ['one', 'two']], names=['first', 'second'])In [302]: indexOut[302]:MultiIndex([(0, 'one'),(0, 'two'),(1, 'one'),(1, 'two'),(2, 'one'),(2, 'two')],names=['first', 'second'])In [303]: index.levels[1]Out[303]: Index(['one', 'two'], dtype='object', name='second')In [304]: index.set_levels(["a", "b"], level=1)Out[304]:MultiIndex([(0, 'a'),(0, 'b'),(1, 'a'),(1, 'b'),(2, 'a'),(2, 'b')],names=['first', 'second'])
Set operations on Index objects
The two main operations are union (|) and intersection (&).
These can be directly called as instance methods or used via overloaded
operators. Difference is provided via the .difference() method.
In [305]: a = pd.Index(['c', 'b', 'a'])In [306]: b = pd.Index(['c', 'e', 'd'])In [307]: a | bOut[307]: Index(['a', 'b', 'c', 'd', 'e'], dtype='object')In [308]: a & bOut[308]: Index(['c'], dtype='object')In [309]: a.difference(b)Out[309]: Index(['a', 'b'], dtype='object')
Also available is the symmetric_difference (^) operation, which returns elements
that appear in either idx1 or idx2, but not in both. This is
equivalent to the Index created by idx1.difference(idx2).union(idx2.difference(idx1)),
with duplicates dropped.
In [310]: idx1 = pd.Index([1, 2, 3, 4])In [311]: idx2 = pd.Index([2, 3, 4, 5])In [312]: idx1.symmetric_difference(idx2)Out[312]: Int64Index([1, 5], dtype='int64')In [313]: idx1 ^ idx2Out[313]: Int64Index([1, 5], dtype='int64')
::: tip Note
The resulting index from a set operation will be sorted in ascending order.
:::
When performing Index.union() between indexes with different dtypes, the indexes
must be cast to a common dtype. Typically, though not always, this is object dtype. The
exception is when performing a union between integer and float data. In this case, the
integer values are converted to float
In [314]: idx1 = pd.Index([0, 1, 2])In [315]: idx2 = pd.Index([0.5, 1.5])In [316]: idx1 | idx2Out[316]: Float64Index([0.0, 0.5, 1.0, 1.5, 2.0], dtype='float64')
Missing values
Even though Index can hold missing values (NaN), it should be avoided
if you do not want any unexpected results. For example, some operations
exclude missing values implicitly.
Index.fillna fills missing values with specified scalar value.
In [317]: idx1 = pd.Index([1, np.nan, 3, 4])In [318]: idx1Out[318]: Float64Index([1.0, nan, 3.0, 4.0], dtype='float64')In [319]: idx1.fillna(2)Out[319]: Float64Index([1.0, 2.0, 3.0, 4.0], dtype='float64')In [320]: idx2 = pd.DatetimeIndex([pd.Timestamp('2011-01-01'),.....: pd.NaT,.....: pd.Timestamp('2011-01-03')]).....:In [321]: idx2Out[321]: DatetimeIndex(['2011-01-01', 'NaT', '2011-01-03'], dtype='datetime64[ns]', freq=None)In [322]: idx2.fillna(pd.Timestamp('2011-01-02'))Out[322]: DatetimeIndex(['2011-01-01', '2011-01-02', '2011-01-03'], dtype='datetime64[ns]', freq=None)
Set / reset index
Occasionally you will load or create a data set into a DataFrame and want to add an index after you’ve already done so. There are a couple of different ways.
Set an index
DataFrame has a set_index() method which takes a column name
(for a regular Index) or a list of column names (for a MultiIndex).
To create a new, re-indexed DataFrame:
In [323]: dataOut[323]:a b c d0 bar one z 1.01 bar two y 2.02 foo one x 3.03 foo two w 4.0In [324]: indexed1 = data.set_index('c')In [325]: indexed1Out[325]:a b dcz bar one 1.0y bar two 2.0x foo one 3.0w foo two 4.0In [326]: indexed2 = data.set_index(['a', 'b'])In [327]: indexed2Out[327]:c da bbar one z 1.0two y 2.0foo one x 3.0two w 4.0
The append keyword option allow you to keep the existing index and append
the given columns to a MultiIndex:
In [328]: frame = data.set_index('c', drop=False)In [329]: frame = frame.set_index(['a', 'b'], append=True)In [330]: frameOut[330]:c dc a bz bar one z 1.0y bar two y 2.0x foo one x 3.0w foo two w 4.0
Other options in set_index allow you not drop the index columns or to add
the index in-place (without creating a new object):
In [331]: data.set_index('c', drop=False)Out[331]:a b c dcz bar one z 1.0y bar two y 2.0x foo one x 3.0w foo two w 4.0In [332]: data.set_index(['a', 'b'], inplace=True)In [333]: dataOut[333]:c da bbar one z 1.0two y 2.0foo one x 3.0two w 4.0
Reset the index
As a convenience, there is a new function on DataFrame called
reset_index() which transfers the index values into the
DataFrame’s columns and sets a simple integer index.
This is the inverse operation of set_index().
In [334]: dataOut[334]:c da bbar one z 1.0two y 2.0foo one x 3.0two w 4.0In [335]: data.reset_index()Out[335]:a b c d0 bar one z 1.01 bar two y 2.02 foo one x 3.03 foo two w 4.0
The output is more similar to a SQL table or a record array. The names for the
columns derived from the index are the ones stored in the names attribute.
You can use the level keyword to remove only a portion of the index:
In [336]: frameOut[336]:c dc a bz bar one z 1.0y bar two y 2.0x foo one x 3.0w foo two w 4.0In [337]: frame.reset_index(level=1)Out[337]:a c dc bz one bar z 1.0y two bar y 2.0x one foo x 3.0w two foo w 4.0
reset_index takes an optional parameter drop which if true simply
discards the index, instead of putting index values in the DataFrame’s columns.
Adding an ad hoc index
If you create an index yourself, you can just assign it to the index field:
data.index = index
Returning a view versus a copy
When setting values in a pandas object, care must be taken to avoid what is called
chained indexing. Here is an example.
In [338]: dfmi = pd.DataFrame([list('abcd'),.....: list('efgh'),.....: list('ijkl'),.....: list('mnop')],.....: columns=pd.MultiIndex.from_product([['one', 'two'],.....: ['first', 'second']])).....:In [339]: dfmiOut[339]:one twofirst second first second0 a b c d1 e f g h2 i j k l3 m n o p
Compare these two access methods:
In [340]: dfmi['one']['second']Out[340]:0 b1 f2 j3 nName: second, dtype: object
In [341]: dfmi.loc[:, ('one', 'second')]Out[341]:0 b1 f2 j3 nName: (one, second), dtype: object
These both yield the same results, so which should you use? It is instructive to understand the order
of operations on these and why method 2 (.loc) is much preferred over method 1 (chained []).
dfmi['one'] selects the first level of the columns and returns a DataFrame that is singly-indexed.
Then another Python operation dfmi_with_one['second'] selects the series indexed by 'second'.
This is indicated by the variable dfmi_with_one because pandas sees these operations as separate events.
e.g. separate calls to __getitem__, so it has to treat them as linear operations, they happen one after another.
Contrast this to df.loc[:,('one','second')] which passes a nested tuple of (slice(None),('one','second')) to a single call to
__getitem__. This allows pandas to deal with this as a single entity. Furthermore this order of operations can be significantly
faster, and allows one to index both axes if so desired.
Why does assignment fail when using chained indexing?
The problem in the previous section is just a performance issue. What’s up with
the SettingWithCopy warning? We don’t usually throw warnings around when
you do something that might cost a few extra milliseconds!
But it turns out that assigning to the product of chained indexing has inherently unpredictable results. To see this, think about how the Python interpreter executes this code:
dfmi.loc[:, ('one', 'second')] = value# becomesdfmi.loc.__setitem__((slice(None), ('one', 'second')), value)
But this code is handled differently:
dfmi['one']['second'] = value# becomesdfmi.__getitem__('one').__setitem__('second', value)
See that __getitem__ in there? Outside of simple cases, it’s very hard to
predict whether it will return a view or a copy (it depends on the memory layout
of the array, about which pandas makes no guarantees), and therefore whether
the __setitem__ will modify dfmi or a temporary object that gets thrown
out immediately afterward. That’s what SettingWithCopy is warning you
about!
::: tip Note
You may be wondering whether we should be concerned about the loc
property in the first example. But dfmi.loc is guaranteed to be dfmi
itself with modified indexing behavior, so dfmi.loc.__getitem__ /
dfmi.loc.__setitem__ operate on dfmi directly. Of course,
dfmi.loc.__getitem__(idx) may be a view or a copy of dfmi.
:::
Sometimes a SettingWithCopy warning will arise at times when there’s no
obvious chained indexing going on. These are the bugs that
SettingWithCopy is designed to catch! Pandas is probably trying to warn you
that you’ve done this:
def do_something(df):foo = df[['bar', 'baz']] # Is foo a view? A copy? Nobody knows!# ... many lines here ...# We don't know whether this will modify df or not!foo['quux'] = valuereturn foo
Yikes!
Evaluation order matters
When you use chained indexing, the order and type of the indexing operation partially determine whether the result is a slice into the original object, or a copy of the slice.
Pandas has the SettingWithCopyWarning because assigning to a copy of a
slice is frequently not intentional, but a mistake caused by chained indexing
returning a copy where a slice was expected.
If you would like pandas to be more or less trusting about assignment to a
chained indexing expression, you can set the option
mode.chained_assignment to one of these values:
'warn', the default, means aSettingWithCopyWarningis printed.'raise'means pandas will raise aSettingWithCopyExceptionyou have to deal with.Nonewill suppress the warnings entirely.
In [342]: dfb = pd.DataFrame({'a': ['one', 'one', 'two',.....: 'three', 'two', 'one', 'six'],.....: 'c': np.arange(7)}).....:# This will show the SettingWithCopyWarning# but the frame values will be setIn [343]: dfb['c'][dfb.a.str.startswith('o')] = 42
This however is operating on a copy and will not work.
>>> pd.set_option('mode.chained_assignment','warn')>>> dfb[dfb.a.str.startswith('o')]['c'] = 42Traceback (most recent call last)...SettingWithCopyWarning:A value is trying to be set on a copy of a slice from a DataFrame.Try using .loc[row_index,col_indexer] = value instead
A chained assignment can also crop up in setting in a mixed dtype frame.
::: tip Note
These setting rules apply to all of .loc/.iloc.
:::
This is the correct access method:
In [344]: dfc = pd.DataFrame({'A': ['aaa', 'bbb', 'ccc'], 'B': [1, 2, 3]})In [345]: dfc.loc[0, 'A'] = 11In [346]: dfcOut[346]:A B0 11 11 bbb 22 ccc 3
This can work at times, but it is not guaranteed to, and therefore should be avoided:
In [347]: dfc = dfc.copy()In [348]: dfc['A'][0] = 111In [349]: dfcOut[349]:A B0 111 11 bbb 22 ccc 3
This will not work at all, and so should be avoided:
>>> pd.set_option('mode.chained_assignment','raise')>>> dfc.loc[0]['A'] = 1111Traceback (most recent call last)...SettingWithCopyException:A value is trying to be set on a copy of a slice from a DataFrame.Try using .loc[row_index,col_indexer] = value instead
::: danger Warning
The chained assignment warnings / exceptions are aiming to inform the user of a possibly invalid assignment. There may be false positives; situations where a chained assignment is inadvertently reported.
:::

