- Group By: split-apply-combine
- Splitting an object into groups
- Iterating through groups
- Selecting a group
- Aggregation
- Transformation
- Filtration
- Dispatching to instance methods
- Flexible
apply - Other useful features
- Automatic exclusion of “nuisance” columns
- Handling of (un)observed Categorical values
- NA and NaT group handling
- Grouping with ordered factors
- Grouping with a grouper specification
- Taking the first rows of each group
- Taking the nth row of each group
- Enumerate group items
- Enumerate groups
- Plotting
- Piping function calls
- Examples
Group By: split-apply-combine
By “group by” we are referring to a process involving one or more of the following steps:
- Splitting the data into groups based on some criteria.
- Applying a function to each group independently.
- Combining the results into a data structure.
Out of these, the split step is the most straightforward. In fact, in many situations we may wish to split the data set into groups and do something with those groups. In the apply step, we might wish to do one of the following:
Aggregation: compute a summary statistic (or statistics) for each group. Some examples:
- Compute group sums or means.
- Compute group sizes / counts.
Transformation: perform some group-specific computations and return a like-indexed object. Some examples:
- Standardize data (zscore) within a group.
- Filling NAs within groups with a value derived from each group.
Filtration: discard some groups, according to a group-wise computation that evaluates True or False. Some examples:
- Discard data that belongs to groups with only a few members.
- Filter out data based on the group sum or mean.
Some combination of the above: GroupBy will examine the results of the apply step and try to return a sensibly combined result if it doesn’t fit into either of the above two categories.
Since the set of object instance methods on pandas data structures are generally
rich and expressive, we often simply want to invoke, say, a DataFrame function
on each group. The name GroupBy should be quite familiar to those who have used
a SQL-based tool (or itertools), in which you can write code like:
SELECT Column1, Column2, mean(Column3), sum(Column4)FROM SomeTableGROUP BY Column1, Column2
We aim to make operations like this natural and easy to express using pandas. We’ll address each area of GroupBy functionality then provide some non-trivial examples / use cases.
See the cookbook for some advanced strategies.
Splitting an object into groups
pandas objects can be split on any of their axes. The abstract definition of grouping is to provide a mapping of labels to group names. To create a GroupBy object (more on what the GroupBy object is later), you may do the following:
In [1]: df = pd.DataFrame([('bird', 'Falconiformes', 389.0),...: ('bird', 'Psittaciformes', 24.0),...: ('mammal', 'Carnivora', 80.2),...: ('mammal', 'Primates', np.nan),...: ('mammal', 'Carnivora', 58)],...: index=['falcon', 'parrot', 'lion', 'monkey', 'leopard'],...: columns=('class', 'order', 'max_speed'))...:In [2]: dfOut[2]:class order max_speedfalcon bird Falconiformes 389.0parrot bird Psittaciformes 24.0lion mammal Carnivora 80.2monkey mammal Primates NaNleopard mammal Carnivora 58.0# default is axis=0In [3]: grouped = df.groupby('class')In [4]: grouped = df.groupby('order', axis='columns')In [5]: grouped = df.groupby(['class', 'order'])
The mapping can be specified many different ways:
- A Python function, to be called on each of the axis labels.
- A list or NumPy array of the same length as the selected axis.
- A dict or
Series, providing alabel -> group namemapping. - For
DataFrameobjects, a string indicating a column to be used to group. Of coursedf.groupby('A')is just syntactic sugar fordf.groupby(df['A']), but it makes life simpler. - For
DataFrameobjects, a string indicating an index level to be used to group. - A list of any of the above things.
Collectively we refer to the grouping objects as the keys. For example,
consider the following DataFrame:
::: tip Note
A string passed to groupby may refer to either a column or an index level.
If a string matches both a column name and an index level name, a
ValueError will be raised.
:::
In [6]: df = pd.DataFrame({'A': ['foo', 'bar', 'foo', 'bar',...: 'foo', 'bar', 'foo', 'foo'],...: 'B': ['one', 'one', 'two', 'three',...: 'two', 'two', 'one', 'three'],...: 'C': np.random.randn(8),...: 'D': np.random.randn(8)})...:In [7]: dfOut[7]:A B C D0 foo one 0.469112 -0.8618491 bar one -0.282863 -2.1045692 foo two -1.509059 -0.4949293 bar three -1.135632 1.0718044 foo two 1.212112 0.7215555 bar two -0.173215 -0.7067716 foo one 0.119209 -1.0395757 foo three -1.044236 0.271860
On a DataFrame, we obtain a GroupBy object by calling groupby().
We could naturally group by either the A or B columns, or both:
In [8]: grouped = df.groupby('A')In [9]: grouped = df.groupby(['A', 'B'])
New in version 0.24.
If we also have a MultiIndex on columns A and B, we can group by all
but the specified columns
In [10]: df2 = df.set_index(['A', 'B'])In [11]: grouped = df2.groupby(level=df2.index.names.difference(['B']))In [12]: grouped.sum()Out[12]:C DAbar -1.591710 -1.739537foo -0.752861 -1.402938
These will split the DataFrame on its index (rows). We could also split by the columns:
In [13]: def get_letter_type(letter):....: if letter.lower() in 'aeiou':....: return 'vowel'....: else:....: return 'consonant'....:In [14]: grouped = df.groupby(get_letter_type, axis=1)
pandas Index objects support duplicate values. If a
non-unique index is used as the group key in a groupby operation, all values
for the same index value will be considered to be in one group and thus the
output of aggregation functions will only contain unique index values:
In [15]: lst = [1, 2, 3, 1, 2, 3]In [16]: s = pd.Series([1, 2, 3, 10, 20, 30], lst)In [17]: grouped = s.groupby(level=0)In [18]: grouped.first()Out[18]:1 12 23 3dtype: int64In [19]: grouped.last()Out[19]:1 102 203 30dtype: int64In [20]: grouped.sum()Out[20]:1 112 223 33dtype: int64
Note that no splitting occurs until it’s needed. Creating the GroupBy object only verifies that you’ve passed a valid mapping.
::: tip Note
Many kinds of complicated data manipulations can be expressed in terms of GroupBy operations (though can’t be guaranteed to be the most efficient). You can get quite creative with the label mapping functions.
:::
GroupBy sorting
By default the group keys are sorted during the groupby operation. You may however pass sort=False for potential speedups:
In [21]: df2 = pd.DataFrame({'X': ['B', 'B', 'A', 'A'], 'Y': [1, 2, 3, 4]})In [22]: df2.groupby(['X']).sum()Out[22]:YXA 7B 3In [23]: df2.groupby(['X'], sort=False).sum()Out[23]:YXB 3A 7
Note that groupby will preserve the order in which observations are sorted within each group.
For example, the groups created by groupby() below are in the order they appeared in the original DataFrame:
In [24]: df3 = pd.DataFrame({'X': ['A', 'B', 'A', 'B'], 'Y': [1, 4, 3, 2]})In [25]: df3.groupby(['X']).get_group('A')Out[25]:X Y0 A 12 A 3In [26]: df3.groupby(['X']).get_group('B')Out[26]:X Y1 B 43 B 2
GroupBy object attributes
The groups attribute is a dict whose keys are the computed unique groups
and corresponding values being the axis labels belonging to each group. In the
above example we have:
In [27]: df.groupby('A').groupsOut[27]:{'bar': Int64Index([1, 3, 5], dtype='int64'),'foo': Int64Index([0, 2, 4, 6, 7], dtype='int64')}In [28]: df.groupby(get_letter_type, axis=1).groupsOut[28]:{'consonant': Index(['B', 'C', 'D'], dtype='object'),'vowel': Index(['A'], dtype='object')}
Calling the standard Python len function on the GroupBy object just returns
the length of the groups dict, so it is largely just a convenience:
In [29]: grouped = df.groupby(['A', 'B'])In [30]: grouped.groupsOut[30]:{('bar', 'one'): Int64Index([1], dtype='int64'),('bar', 'three'): Int64Index([3], dtype='int64'),('bar', 'two'): Int64Index([5], dtype='int64'),('foo', 'one'): Int64Index([0, 6], dtype='int64'),('foo', 'three'): Int64Index([7], dtype='int64'),('foo', 'two'): Int64Index([2, 4], dtype='int64')}In [31]: len(grouped)Out[31]: 6
GroupBy will tab complete column names (and other attributes):
In [32]: dfOut[32]:height weight gender2000-01-01 42.849980 157.500553 male2000-01-02 49.607315 177.340407 male2000-01-03 56.293531 171.524640 male2000-01-04 48.421077 144.251986 female2000-01-05 46.556882 152.526206 male2000-01-06 68.448851 168.272968 female2000-01-07 70.757698 136.431469 male2000-01-08 58.909500 176.499753 female2000-01-09 76.435631 174.094104 female2000-01-10 45.306120 177.540920 maleIn [33]: gb = df.groupby('gender')
In [34]: gb.<TAB> # noqa: E225, E999gb.agg gb.boxplot gb.cummin gb.describe gb.filter gb.get_group gb.height gb.last gb.median gb.ngroups gb.plot gb.rank gb.std gb.transformgb.aggregate gb.count gb.cumprod gb.dtype gb.first gb.groups gb.hist gb.max gb.min gb.nth gb.prod gb.resample gb.sum gb.vargb.apply gb.cummax gb.cumsum gb.fillna gb.gender gb.head gb.indices gb.mean gb.name gb.ohlc gb.quantile gb.size gb.tail gb.weight
GroupBy with MultiIndex
With hierarchically-indexed data, it’s quite natural to group by one of the levels of the hierarchy.
Let’s create a Series with a two-level MultiIndex.
In [35]: arrays = [['bar', 'bar', 'baz', 'baz', 'foo', 'foo', 'qux', 'qux'],....: ['one', 'two', 'one', 'two', 'one', 'two', 'one', 'two']]....:In [36]: index = pd.MultiIndex.from_arrays(arrays, names=['first', 'second'])In [37]: s = pd.Series(np.random.randn(8), index=index)In [38]: sOut[38]:first secondbar one -0.919854two -0.042379baz one 1.247642two -0.009920foo one 0.290213two 0.495767qux one 0.362949two 1.548106dtype: float64
We can then group by one of the levels in s.
In [39]: grouped = s.groupby(level=0)In [40]: grouped.sum()Out[40]:firstbar -0.962232baz 1.237723foo 0.785980qux 1.911055dtype: float64
If the MultiIndex has names specified, these can be passed instead of the level number:
In [41]: s.groupby(level='second').sum()Out[41]:secondone 0.980950two 1.991575dtype: float64
The aggregation functions such as sum will take the level parameter
directly. Additionally, the resulting index will be named according to the
chosen level:
In [42]: s.sum(level='second')Out[42]:secondone 0.980950two 1.991575dtype: float64
Grouping with multiple levels is supported.
In [43]: sOut[43]:first second thirdbar doo one -1.131345two -0.089329baz bee one 0.337863two -0.945867foo bop one -0.932132two 1.956030qux bop one 0.017587two -0.016692dtype: float64In [44]: s.groupby(level=['first', 'second']).sum()Out[44]:first secondbar doo -1.220674baz bee -0.608004foo bop 1.023898qux bop 0.000895dtype: float64
New in version 0.20.
Index level names may be supplied as keys.
In [45]: s.groupby(['first', 'second']).sum()Out[45]:first secondbar doo -1.220674baz bee -0.608004foo bop 1.023898qux bop 0.000895dtype: float64
More on the sum function and aggregation later.
Grouping DataFrame with Index levels and columns
A DataFrame may be grouped by a combination of columns and index levels by
specifying the column names as strings and the index levels as pd.Grouper
objects.
In [46]: arrays = [['bar', 'bar', 'baz', 'baz', 'foo', 'foo', 'qux', 'qux'],....: ['one', 'two', 'one', 'two', 'one', 'two', 'one', 'two']]....:In [47]: index = pd.MultiIndex.from_arrays(arrays, names=['first', 'second'])In [48]: df = pd.DataFrame({'A': [1, 1, 1, 1, 2, 2, 3, 3],....: 'B': np.arange(8)},....: index=index)....:In [49]: dfOut[49]:A Bfirst secondbar one 1 0two 1 1baz one 1 2two 1 3foo one 2 4two 2 5qux one 3 6two 3 7
The following example groups df by the second index level and
the A column.
In [50]: df.groupby([pd.Grouper(level=1), 'A']).sum()Out[50]:Bsecond Aone 1 22 43 6two 1 42 53 7
Index levels may also be specified by name.
In [51]: df.groupby([pd.Grouper(level='second'), 'A']).sum()Out[51]:Bsecond Aone 1 22 43 6two 1 42 53 7
New in version 0.20.
Index level names may be specified as keys directly to groupby.
In [52]: df.groupby(['second', 'A']).sum()Out[52]:Bsecond Aone 1 22 43 6two 1 42 53 7
DataFrame column selection in GroupBy
Once you have created the GroupBy object from a DataFrame, you might want to do
something different for each of the columns. Thus, using [] similar to
getting a column from a DataFrame, you can do:
In [53]: grouped = df.groupby(['A'])In [54]: grouped_C = grouped['C']In [55]: grouped_D = grouped['D']
This is mainly syntactic sugar for the alternative and much more verbose:
In [56]: df['C'].groupby(df['A'])Out[56]: <pandas.core.groupby.generic.SeriesGroupBy object at 0x7f65f21ac518>
Additionally this method avoids recomputing the internal grouping information derived from the passed key.
Iterating through groups
With the GroupBy object in hand, iterating through the grouped data is very
natural and functions similarly to itertools.groupby():
In [57]: grouped = df.groupby('A')In [58]: for name, group in grouped:....: print(name)....: print(group)....:barA B C D1 bar one 0.254161 1.5117633 bar three 0.215897 -0.9905825 bar two -0.077118 1.211526fooA B C D0 foo one -0.575247 1.3460612 foo two -1.143704 1.6270814 foo two 1.193555 -0.4416526 foo one -0.408530 0.2685207 foo three -0.862495 0.024580
In the case of grouping by multiple keys, the group name will be a tuple:
In [59]: for name, group in df.groupby(['A', 'B']):....: print(name)....: print(group)....:('bar', 'one')A B C D1 bar one 0.254161 1.511763('bar', 'three')A B C D3 bar three 0.215897 -0.990582('bar', 'two')A B C D5 bar two -0.077118 1.211526('foo', 'one')A B C D0 foo one -0.575247 1.3460616 foo one -0.408530 0.268520('foo', 'three')A B C D7 foo three -0.862495 0.02458('foo', 'two')A B C D2 foo two -1.143704 1.6270814 foo two 1.193555 -0.441652
Selecting a group
A single group can be selected using
get_group():
In [60]: grouped.get_group('bar')Out[60]:A B C D1 bar one 0.254161 1.5117633 bar three 0.215897 -0.9905825 bar two -0.077118 1.211526
Or for an object grouped on multiple columns:
In [61]: df.groupby(['A', 'B']).get_group(('bar', 'one'))Out[61]:A B C D1 bar one 0.254161 1.511763
Aggregation
Once the GroupBy object has been created, several methods are available to perform a computation on the grouped data. These operations are similar to the aggregating API, window functions API, and resample API.
An obvious one is aggregation via the
aggregate() or equivalently
agg() method:
In [62]: grouped = df.groupby('A')In [63]: grouped.aggregate(np.sum)Out[63]:C DAbar 0.392940 1.732707foo -1.796421 2.824590In [64]: grouped = df.groupby(['A', 'B'])In [65]: grouped.aggregate(np.sum)Out[65]:C DA Bbar one 0.254161 1.511763three 0.215897 -0.990582two -0.077118 1.211526foo one -0.983776 1.614581three -0.862495 0.024580two 0.049851 1.185429
As you can see, the result of the aggregation will have the group names as the
new index along the grouped axis. In the case of multiple keys, the result is a
MultiIndex by default, though this can be
changed by using the as_index option:
In [66]: grouped = df.groupby(['A', 'B'], as_index=False)In [67]: grouped.aggregate(np.sum)Out[67]:A B C D0 bar one 0.254161 1.5117631 bar three 0.215897 -0.9905822 bar two -0.077118 1.2115263 foo one -0.983776 1.6145814 foo three -0.862495 0.0245805 foo two 0.049851 1.185429In [68]: df.groupby('A', as_index=False).sum()Out[68]:A C D0 bar 0.392940 1.7327071 foo -1.796421 2.824590
Note that you could use the reset_index DataFrame function to achieve the
same result as the column names are stored in the resulting MultiIndex:
In [69]: df.groupby(['A', 'B']).sum().reset_index()Out[69]:A B C D0 bar one 0.254161 1.5117631 bar three 0.215897 -0.9905822 bar two -0.077118 1.2115263 foo one -0.983776 1.6145814 foo three -0.862495 0.0245805 foo two 0.049851 1.185429
Another simple aggregation example is to compute the size of each group.
This is included in GroupBy as the size method. It returns a Series whose
index are the group names and whose values are the sizes of each group.
In [70]: grouped.size()Out[70]:A Bbar one 1three 1two 1foo one 2three 1two 2dtype: int64
In [71]: grouped.describe()Out[71]:C Dcount mean std min 25% 50% 75% max count mean std min 25% 50% 75% max0 1.0 0.254161 NaN 0.254161 0.254161 0.254161 0.254161 0.254161 1.0 1.511763 NaN 1.511763 1.511763 1.511763 1.511763 1.5117631 1.0 0.215897 NaN 0.215897 0.215897 0.215897 0.215897 0.215897 1.0 -0.990582 NaN -0.990582 -0.990582 -0.990582 -0.990582 -0.9905822 1.0 -0.077118 NaN -0.077118 -0.077118 -0.077118 -0.077118 -0.077118 1.0 1.211526 NaN 1.211526 1.211526 1.211526 1.211526 1.2115263 2.0 -0.491888 0.117887 -0.575247 -0.533567 -0.491888 -0.450209 -0.408530 2.0 0.807291 0.761937 0.268520 0.537905 0.807291 1.076676 1.3460614 1.0 -0.862495 NaN -0.862495 -0.862495 -0.862495 -0.862495 -0.862495 1.0 0.024580 NaN 0.024580 0.024580 0.024580 0.024580 0.0245805 2.0 0.024925 1.652692 -1.143704 -0.559389 0.024925 0.609240 1.193555 2.0 0.592714 1.462816 -0.441652 0.075531 0.592714 1.109898 1.627081
::: tip Note
Aggregation functions will not return the groups that you are aggregating over
if they are named columns, when as_index=True, the default. The grouped columns will
be the indices of the returned object.
Passing as_index=False will return the groups that you are aggregating over, if they are
named columns.
:::
Aggregating functions are the ones that reduce the dimension of the returned objects. Some common aggregating functions are tabulated below:
| Function | Description |
|---|---|
| mean() | Compute mean of groups |
| sum() | Compute sum of group values |
| size() | Compute group sizes |
| count() | Compute count of group |
| std() | Standard deviation of groups |
| var() | Compute variance of groups |
| sem() | Standard error of the mean of groups |
| describe() | Generates descriptive statistics |
| first() | Compute first of group values |
| last() | Compute last of group values |
| nth() | Take nth value, or a subset if n is a list |
| min() | Compute min of group values |
| max() | Compute max of group values |
The aggregating functions above will exclude NA values. Any function which
reduces a Series to a scalar value is an aggregation function and will work,
a trivial example is df.groupby('A').agg(lambda ser: 1). Note that
nth() can act as a reducer or a
filter, see here.
Applying multiple functions at once
With grouped Series you can also pass a list or dict of functions to do
aggregation with, outputting a DataFrame:
In [72]: grouped = df.groupby('A')In [73]: grouped['C'].agg([np.sum, np.mean, np.std])Out[73]:sum mean stdAbar 0.392940 0.130980 0.181231foo -1.796421 -0.359284 0.912265
On a grouped DataFrame, you can pass a list of functions to apply to each
column, which produces an aggregated result with a hierarchical index:
In [74]: grouped.agg([np.sum, np.mean, np.std])Out[74]:C Dsum mean std sum mean stdAbar 0.392940 0.130980 0.181231 1.732707 0.577569 1.366330foo -1.796421 -0.359284 0.912265 2.824590 0.564918 0.884785
The resulting aggregations are named for the functions themselves. If you
need to rename, then you can add in a chained operation for a Series like this:
In [75]: (grouped['C'].agg([np.sum, np.mean, np.std])....: .rename(columns={'sum': 'foo',....: 'mean': 'bar',....: 'std': 'baz'}))....:Out[75]:foo bar bazAbar 0.392940 0.130980 0.181231foo -1.796421 -0.359284 0.912265
For a grouped DataFrame, you can rename in a similar manner:
In [76]: (grouped.agg([np.sum, np.mean, np.std])....: .rename(columns={'sum': 'foo',....: 'mean': 'bar',....: 'std': 'baz'}))....:Out[76]:C Dfoo bar baz foo bar bazAbar 0.392940 0.130980 0.181231 1.732707 0.577569 1.366330foo -1.796421 -0.359284 0.912265 2.824590 0.564918 0.884785
::: tip Note
In general, the output column names should be unique. You can’t apply the same function (or two functions with the same name) to the same column.
In [77]: grouped['C'].agg(['sum', 'sum'])---------------------------------------------------------------------------SpecificationError Traceback (most recent call last)<ipython-input-77-7be02859f395> in <module>----> 1 grouped['C'].agg(['sum', 'sum'])/pandas/pandas/core/groupby/generic.py in aggregate(self, func_or_funcs, *args, **kwargs)849 # but not the class list / tuple itself.850 func_or_funcs = _maybe_mangle_lambdas(func_or_funcs)--> 851 ret = self._aggregate_multiple_funcs(func_or_funcs, (_level or 0) + 1)852 if relabeling:853 ret.columns = columns/pandas/pandas/core/groupby/generic.py in _aggregate_multiple_funcs(self, arg, _level)919 raise SpecificationError(920 "Function names must be unique, found multiple named "--> 921 "{}".format(name)922 )923SpecificationError: Function names must be unique, found multiple named sum
Pandas does allow you to provide multiple lambdas. In this case, pandas
will mangle the name of the (nameless) lambda functions, appending _
to each subsequent lambda.
In [78]: grouped['C'].agg([lambda x: x.max() - x.min(),....: lambda x: x.median() - x.mean()])....:Out[78]:<lambda_0> <lambda_1>Abar 0.331279 0.084917foo 2.337259 -0.215962
:::
Named aggregation
New in version 0.25.0.
To support column-specific aggregation with control over the output column names, pandas
accepts the special syntax in GroupBy.agg(), known as “named aggregation”, where
- The keywords are the output column names
- The values are tuples whose first element is the column to select
and the second element is the aggregation to apply to that column. Pandas
provides the
pandas.NamedAggnamedtuple with the fields['column', 'aggfunc']to make it clearer what the arguments are. As usual, the aggregation can be a callable or a string alias.
In [79]: animals = pd.DataFrame({'kind': ['cat', 'dog', 'cat', 'dog'],....: 'height': [9.1, 6.0, 9.5, 34.0],....: 'weight': [7.9, 7.5, 9.9, 198.0]})....:In [80]: animalsOut[80]:kind height weight0 cat 9.1 7.91 dog 6.0 7.52 cat 9.5 9.93 dog 34.0 198.0In [81]: animals.groupby("kind").agg(....: min_height=pd.NamedAgg(column='height', aggfunc='min'),....: max_height=pd.NamedAgg(column='height', aggfunc='max'),....: average_weight=pd.NamedAgg(column='weight', aggfunc=np.mean),....: )....:Out[81]:min_height max_height average_weightkindcat 9.1 9.5 8.90dog 6.0 34.0 102.75
pandas.NamedAgg is just a namedtuple. Plain tuples are allowed as well.
In [82]: animals.groupby("kind").agg(....: min_height=('height', 'min'),....: max_height=('height', 'max'),....: average_weight=('weight', np.mean),....: )....:Out[82]:min_height max_height average_weightkindcat 9.1 9.5 8.90dog 6.0 34.0 102.75
If your desired output column names are not valid python keywords, construct a dictionary and unpack the keyword arguments
In [83]: animals.groupby("kind").agg(**{....: 'total weight': pd.NamedAgg(column='weight', aggfunc=sum),....: })....:Out[83]:total weightkindcat 17.8dog 205.5
Additional keyword arguments are not passed through to the aggregation functions. Only pairs
of (column, aggfunc) should be passed as **kwargs. If your aggregation functions
requires additional arguments, partially apply them with functools.partial().
::: tip Note
For Python 3.5 and earlier, the order of **kwargs in a functions was not
preserved. This means that the output column ordering would not be
consistent. To ensure consistent ordering, the keys (and so output columns)
will always be sorted for Python 3.5.
:::
Named aggregation is also valid for Series groupby aggregations. In this case there’s no column selection, so the values are just the functions.
In [84]: animals.groupby("kind").height.agg(....: min_height='min',....: max_height='max',....: )....:Out[84]:min_height max_heightkindcat 9.1 9.5dog 6.0 34.0
Applying different functions to DataFrame columns
By passing a dict to aggregate you can apply a different aggregation to the
columns of a DataFrame:
In [85]: grouped.agg({'C': np.sum,....: 'D': lambda x: np.std(x, ddof=1)})....:Out[85]:C DAbar 0.392940 1.366330foo -1.796421 0.884785
The function names can also be strings. In order for a string to be valid it must be either implemented on GroupBy or available via dispatching:
In [86]: grouped.agg({'C': 'sum', 'D': 'std'})Out[86]:C DAbar 0.392940 1.366330foo -1.796421 0.884785
Cython-optimized aggregation functions
Some common aggregations, currently only sum, mean, std, and sem, have
optimized Cython implementations:
In [87]: df.groupby('A').sum()Out[87]:C DAbar 0.392940 1.732707foo -1.796421 2.824590In [88]: df.groupby(['A', 'B']).mean()Out[88]:C DA Bbar one 0.254161 1.511763three 0.215897 -0.990582two -0.077118 1.211526foo one -0.491888 0.807291three -0.862495 0.024580two 0.024925 0.592714
Of course sum and mean are implemented on pandas objects, so the above
code would work even without the special versions via dispatching (see below).
Transformation
The transform method returns an object that is indexed the same (same size)
as the one being grouped. The transform function must:
- Return a result that is either the same size as the group chunk or
broadcastable to the size of the group chunk (e.g., a scalar,
grouped.transform(lambda x: x.iloc[-1])). - Operate column-by-column on the group chunk. The transform is applied to the first group chunk using chunk.apply.
- Not perform in-place operations on the group chunk. Group chunks should
be treated as immutable, and changes to a group chunk may produce unexpected
results. For example, when using
fillna,inplacemust beFalse(grouped.transform(lambda x: x.fillna(inplace=False))). - (Optionally) operates on the entire group chunk. If this is supported, a fast path is used starting from the second chunk.
For example, suppose we wished to standardize the data within each group:
In [89]: index = pd.date_range('10/1/1999', periods=1100)In [90]: ts = pd.Series(np.random.normal(0.5, 2, 1100), index)In [91]: ts = ts.rolling(window=100, min_periods=100).mean().dropna()In [92]: ts.head()Out[92]:2000-01-08 0.7793332000-01-09 0.7788522000-01-10 0.7864762000-01-11 0.7827972000-01-12 0.798110Freq: D, dtype: float64In [93]: ts.tail()Out[93]:2002-09-30 0.6602942002-10-01 0.6310952002-10-02 0.6736012002-10-03 0.7092132002-10-04 0.719369Freq: D, dtype: float64In [94]: transformed = (ts.groupby(lambda x: x.year)....: .transform(lambda x: (x - x.mean()) / x.std()))....:
We would expect the result to now have mean 0 and standard deviation 1 within each group, which we can easily check:
# Original DataIn [95]: grouped = ts.groupby(lambda x: x.year)In [96]: grouped.mean()Out[96]:2000 0.4424412001 0.5262462002 0.459365dtype: float64In [97]: grouped.std()Out[97]:2000 0.1317522001 0.2109452002 0.128753dtype: float64# Transformed DataIn [98]: grouped_trans = transformed.groupby(lambda x: x.year)In [99]: grouped_trans.mean()Out[99]:2000 1.168208e-152001 1.454544e-152002 1.726657e-15dtype: float64In [100]: grouped_trans.std()Out[100]:2000 1.02001 1.02002 1.0dtype: float64
We can also visually compare the original and transformed data sets.
In [101]: compare = pd.DataFrame({'Original': ts, 'Transformed': transformed})In [102]: compare.plot()Out[102]: <matplotlib.axes._subplots.AxesSubplot at 0x7f65f731cba8>
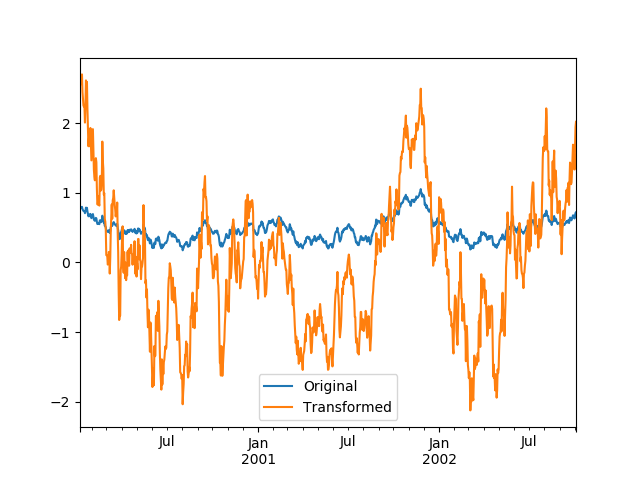
Transformation functions that have lower dimension outputs are broadcast to match the shape of the input array.
In [103]: ts.groupby(lambda x: x.year).transform(lambda x: x.max() - x.min())Out[103]:2000-01-08 0.6238932000-01-09 0.6238932000-01-10 0.6238932000-01-11 0.6238932000-01-12 0.623893...2002-09-30 0.5582752002-10-01 0.5582752002-10-02 0.5582752002-10-03 0.5582752002-10-04 0.558275Freq: D, Length: 1001, dtype: float64
Alternatively, the built-in methods could be used to produce the same outputs.
In [104]: max = ts.groupby(lambda x: x.year).transform('max')In [105]: min = ts.groupby(lambda x: x.year).transform('min')In [106]: max - minOut[106]:2000-01-08 0.6238932000-01-09 0.6238932000-01-10 0.6238932000-01-11 0.6238932000-01-12 0.623893...2002-09-30 0.5582752002-10-01 0.5582752002-10-02 0.5582752002-10-03 0.5582752002-10-04 0.558275Freq: D, Length: 1001, dtype: float64
Another common data transform is to replace missing data with the group mean.
In [107]: data_dfOut[107]:A B C0 1.539708 -1.166480 0.5330261 1.302092 -0.505754 NaN2 -0.371983 1.104803 -0.6515203 -1.309622 1.118697 -1.1616574 -1.924296 0.396437 0.812436.. ... ... ...995 -0.093110 0.683847 -0.774753996 -0.185043 1.438572 NaN997 -0.394469 -0.642343 0.011374998 -1.174126 1.857148 NaN999 0.234564 0.517098 0.393534[1000 rows x 3 columns]In [108]: countries = np.array(['US', 'UK', 'GR', 'JP'])In [109]: key = countries[np.random.randint(0, 4, 1000)]In [110]: grouped = data_df.groupby(key)# Non-NA count in each groupIn [111]: grouped.count()Out[111]:A B CGR 209 217 189JP 240 255 217UK 216 231 193US 239 250 217In [112]: transformed = grouped.transform(lambda x: x.fillna(x.mean()))
We can verify that the group means have not changed in the transformed data and that the transformed data contains no NAs.
In [113]: grouped_trans = transformed.groupby(key)In [114]: grouped.mean() # original group meansOut[114]:A B CGR -0.098371 -0.015420 0.068053JP 0.069025 0.023100 -0.077324UK 0.034069 -0.052580 -0.116525US 0.058664 -0.020399 0.028603In [115]: grouped_trans.mean() # transformation did not change group meansOut[115]:A B CGR -0.098371 -0.015420 0.068053JP 0.069025 0.023100 -0.077324UK 0.034069 -0.052580 -0.116525US 0.058664 -0.020399 0.028603In [116]: grouped.count() # original has some missing data pointsOut[116]:A B CGR 209 217 189JP 240 255 217UK 216 231 193US 239 250 217In [117]: grouped_trans.count() # counts after transformationOut[117]:A B CGR 228 228 228JP 267 267 267UK 247 247 247US 258 258 258In [118]: grouped_trans.size() # Verify non-NA count equals group sizeOut[118]:GR 228JP 267UK 247US 258dtype: int64
::: tip Note
Some functions will automatically transform the input when applied to a
GroupBy object, but returning an object of the same shape as the original.
Passing as_index=False will not affect these transformation methods.
For example: fillna, ffill, bfill, shift..
In [119]: grouped.ffill()Out[119]:A B C0 1.539708 -1.166480 0.5330261 1.302092 -0.505754 0.5330262 -0.371983 1.104803 -0.6515203 -1.309622 1.118697 -1.1616574 -1.924296 0.396437 0.812436.. ... ... ...995 -0.093110 0.683847 -0.774753996 -0.185043 1.438572 -0.774753997 -0.394469 -0.642343 0.011374998 -1.174126 1.857148 -0.774753999 0.234564 0.517098 0.393534[1000 rows x 3 columns]
:::
New syntax to window and resample operations
New in version 0.18.1.
Working with the resample, expanding or rolling operations on the groupby
level used to require the application of helper functions. However,
now it is possible to use resample(), expanding() and
rolling() as methods on groupbys.
The example below will apply the rolling() method on the samples of
the column B based on the groups of column A.
In [120]: df_re = pd.DataFrame({'A': [1] * 10 + [5] * 10,.....: 'B': np.arange(20)}).....:In [121]: df_reOut[121]:A B0 1 01 1 12 1 23 1 34 1 4.. .. ..15 5 1516 5 1617 5 1718 5 1819 5 19[20 rows x 2 columns]In [122]: df_re.groupby('A').rolling(4).B.mean()Out[122]:A1 0 NaN1 NaN2 NaN3 1.54 2.5...5 15 13.516 14.517 15.518 16.519 17.5Name: B, Length: 20, dtype: float64
The expanding() method will accumulate a given operation
(sum() in the example) for all the members of each particular
group.
In [123]: df_re.groupby('A').expanding().sum()Out[123]:A BA1 0 1.0 0.01 2.0 1.02 3.0 3.03 4.0 6.04 5.0 10.0... ... ...5 15 30.0 75.016 35.0 91.017 40.0 108.018 45.0 126.019 50.0 145.0[20 rows x 2 columns]
Suppose you want to use the resample() method to get a daily
frequency in each group of your dataframe and wish to complete the
missing values with the ffill() method.
In [124]: df_re = pd.DataFrame({'date': pd.date_range(start='2016-01-01', periods=4,.....: freq='W'),.....: 'group': [1, 1, 2, 2],.....: 'val': [5, 6, 7, 8]}).set_index('date').....:In [125]: df_reOut[125]:group valdate2016-01-03 1 52016-01-10 1 62016-01-17 2 72016-01-24 2 8In [126]: df_re.groupby('group').resample('1D').ffill()Out[126]:group valgroup date1 2016-01-03 1 52016-01-04 1 52016-01-05 1 52016-01-06 1 52016-01-07 1 5... ... ...2 2016-01-20 2 72016-01-21 2 72016-01-22 2 72016-01-23 2 72016-01-24 2 8[16 rows x 2 columns]
Filtration
The filter method returns a subset of the original object. Suppose we
want to take only elements that belong to groups with a group sum greater
than 2.
In [127]: sf = pd.Series([1, 1, 2, 3, 3, 3])In [128]: sf.groupby(sf).filter(lambda x: x.sum() > 2)Out[128]:3 34 35 3dtype: int64
The argument of filter must be a function that, applied to the group as a
whole, returns True or False.
Another useful operation is filtering out elements that belong to groups with only a couple members.
In [129]: dff = pd.DataFrame({'A': np.arange(8), 'B': list('aabbbbcc')})In [130]: dff.groupby('B').filter(lambda x: len(x) > 2)Out[130]:A B2 2 b3 3 b4 4 b5 5 b
Alternatively, instead of dropping the offending groups, we can return a like-indexed objects where the groups that do not pass the filter are filled with NaNs.
In [131]: dff.groupby('B').filter(lambda x: len(x) > 2, dropna=False)Out[131]:A B0 NaN NaN1 NaN NaN2 2.0 b3 3.0 b4 4.0 b5 5.0 b6 NaN NaN7 NaN NaN
For DataFrames with multiple columns, filters should explicitly specify a column as the filter criterion.
In [132]: dff['C'] = np.arange(8)In [133]: dff.groupby('B').filter(lambda x: len(x['C']) > 2)Out[133]:A B C2 2 b 23 3 b 34 4 b 45 5 b 5
::: tip Note
Some functions when applied to a groupby object will act as a filter on the input, returning
a reduced shape of the original (and potentially eliminating groups), but with the index unchanged.
Passing as_index=False will not affect these transformation methods.
For example: head, tail.
In [134]: dff.groupby('B').head(2)Out[134]:A B C0 0 a 01 1 a 12 2 b 23 3 b 36 6 c 67 7 c 7
:::
Dispatching to instance methods
When doing an aggregation or transformation, you might just want to call an instance method on each data group. This is pretty easy to do by passing lambda functions:
In [135]: grouped = df.groupby('A')In [136]: grouped.agg(lambda x: x.std())Out[136]:C DAbar 0.181231 1.366330foo 0.912265 0.884785
But, it’s rather verbose and can be untidy if you need to pass additional arguments. Using a bit of metaprogramming cleverness, GroupBy now has the ability to “dispatch” method calls to the groups:
In [137]: grouped.std()Out[137]:C DAbar 0.181231 1.366330foo 0.912265 0.884785
What is actually happening here is that a function wrapper is being
generated. When invoked, it takes any passed arguments and invokes the function
with any arguments on each group (in the above example, the std
function). The results are then combined together much in the style of agg
and transform (it actually uses apply to infer the gluing, documented
next). This enables some operations to be carried out rather succinctly:
In [138]: tsdf = pd.DataFrame(np.random.randn(1000, 3),.....: index=pd.date_range('1/1/2000', periods=1000),.....: columns=['A', 'B', 'C']).....:In [139]: tsdf.iloc[::2] = np.nanIn [140]: grouped = tsdf.groupby(lambda x: x.year)In [141]: grouped.fillna(method='pad')Out[141]:A B C2000-01-01 NaN NaN NaN2000-01-02 -0.353501 -0.080957 -0.8768642000-01-03 -0.353501 -0.080957 -0.8768642000-01-04 0.050976 0.044273 -0.5598492000-01-05 0.050976 0.044273 -0.559849... ... ... ...2002-09-22 0.005011 0.053897 -1.0269222002-09-23 0.005011 0.053897 -1.0269222002-09-24 -0.456542 -1.849051 1.5598562002-09-25 -0.456542 -1.849051 1.5598562002-09-26 1.123162 0.354660 1.128135[1000 rows x 3 columns]
In this example, we chopped the collection of time series into yearly chunks then independently called fillna on the groups.
The nlargest and nsmallest methods work on Series style groupbys:
In [142]: s = pd.Series([9, 8, 7, 5, 19, 1, 4.2, 3.3])In [143]: g = pd.Series(list('abababab'))In [144]: gb = s.groupby(g)In [145]: gb.nlargest(3)Out[145]:a 4 19.00 9.02 7.0b 1 8.03 5.07 3.3dtype: float64In [146]: gb.nsmallest(3)Out[146]:a 6 4.22 7.00 9.0b 5 1.07 3.33 5.0dtype: float64
Flexible apply
Some operations on the grouped data might not fit into either the aggregate or
transform categories. Or, you may simply want GroupBy to infer how to combine
the results. For these, use the apply function, which can be substituted
for both aggregate and transform in many standard use cases. However,
apply can handle some exceptional use cases, for example:
In [147]: dfOut[147]:A B C D0 foo one -0.575247 1.3460611 bar one 0.254161 1.5117632 foo two -1.143704 1.6270813 bar three 0.215897 -0.9905824 foo two 1.193555 -0.4416525 bar two -0.077118 1.2115266 foo one -0.408530 0.2685207 foo three -0.862495 0.024580In [148]: grouped = df.groupby('A')# could also just call .describe()In [149]: grouped['C'].apply(lambda x: x.describe())Out[149]:Abar count 3.000000mean 0.130980std 0.181231min -0.07711825% 0.069390...foo min -1.14370425% -0.86249550% -0.57524775% -0.408530max 1.193555Name: C, Length: 16, dtype: float64
The dimension of the returned result can also change:
In [150]: grouped = df.groupby('A')['C']In [151]: def f(group):.....: return pd.DataFrame({'original': group,.....: 'demeaned': group - group.mean()}).....:In [152]: grouped.apply(f)Out[152]:original demeaned0 -0.575247 -0.2159621 0.254161 0.1231812 -1.143704 -0.7844203 0.215897 0.0849174 1.193555 1.5528395 -0.077118 -0.2080986 -0.408530 -0.0492457 -0.862495 -0.503211
apply on a Series can operate on a returned value from the applied function,
that is itself a series, and possibly upcast the result to a DataFrame:
In [153]: def f(x):.....: return pd.Series([x, x ** 2], index=['x', 'x^2']).....:In [154]: s = pd.Series(np.random.rand(5))In [155]: sOut[155]:0 0.3214381 0.4934962 0.1395053 0.9101034 0.194158dtype: float64In [156]: s.apply(f)Out[156]:x x^20 0.321438 0.1033231 0.493496 0.2435382 0.139505 0.0194623 0.910103 0.8282874 0.194158 0.037697
::: tip Note
apply can act as a reducer, transformer, or filter function, depending on exactly what is passed to it.
So depending on the path taken, and exactly what you are grouping. Thus the grouped columns(s) may be included in
the output as well as set the indices.
:::
Other useful features
Automatic exclusion of “nuisance” columns
Again consider the example DataFrame we’ve been looking at:
In [157]: dfOut[157]:A B C D0 foo one -0.575247 1.3460611 bar one 0.254161 1.5117632 foo two -1.143704 1.6270813 bar three 0.215897 -0.9905824 foo two 1.193555 -0.4416525 bar two -0.077118 1.2115266 foo one -0.408530 0.2685207 foo three -0.862495 0.024580
Suppose we wish to compute the standard deviation grouped by the A
column. There is a slight problem, namely that we don’t care about the data in
column B. We refer to this as a “nuisance” column. If the passed
aggregation function can’t be applied to some columns, the troublesome columns
will be (silently) dropped. Thus, this does not pose any problems:
In [158]: df.groupby('A').std()Out[158]:C DAbar 0.181231 1.366330foo 0.912265 0.884785
Note that df.groupby('A').colname.std(). is more efficient than
df.groupby('A').std().colname, so if the result of an aggregation function
is only interesting over one column (here colname), it may be filtered
before applying the aggregation function.
::: tip Note
Any object column, also if it contains numerical values such as Decimal
objects, is considered as a “nuisance” columns. They are excluded from
aggregate functions automatically in groupby.
If you do wish to include decimal or object columns in an aggregation with other non-nuisance data types, you must do so explicitly.
:::
In [159]: from decimal import DecimalIn [160]: df_dec = pd.DataFrame(.....: {'id': [1, 2, 1, 2],.....: 'int_column': [1, 2, 3, 4],.....: 'dec_column': [Decimal('0.50'), Decimal('0.15'),.....: Decimal('0.25'), Decimal('0.40')].....: }.....: ).....:# Decimal columns can be sum'd explicitly by themselves...In [161]: df_dec.groupby(['id'])[['dec_column']].sum()Out[161]:dec_columnid1 0.752 0.55# ...but cannot be combined with standard data types or they will be excludedIn [162]: df_dec.groupby(['id'])[['int_column', 'dec_column']].sum()Out[162]:int_columnid1 42 6# Use .agg function to aggregate over standard and "nuisance" data types# at the same timeIn [163]: df_dec.groupby(['id']).agg({'int_column': 'sum', 'dec_column': 'sum'})Out[163]:int_column dec_columnid1 4 0.752 6 0.55
Handling of (un)observed Categorical values
When using a Categorical grouper (as a single grouper, or as part of multiple groupers), the observed keyword
controls whether to return a cartesian product of all possible groupers values (observed=False) or only those
that are observed groupers (observed=True).
Show all values:
In [164]: pd.Series([1, 1, 1]).groupby(pd.Categorical(['a', 'a', 'a'],.....: categories=['a', 'b']),.....: observed=False).count().....:Out[164]:a 3b 0dtype: int64
Show only the observed values:
In [165]: pd.Series([1, 1, 1]).groupby(pd.Categorical(['a', 'a', 'a'],.....: categories=['a', 'b']),.....: observed=True).count().....:Out[165]:a 3dtype: int64
The returned dtype of the grouped will always include all of the categories that were grouped.
In [166]: s = pd.Series([1, 1, 1]).groupby(pd.Categorical(['a', 'a', 'a'],.....: categories=['a', 'b']),.....: observed=False).count().....:In [167]: s.index.dtypeOut[167]: CategoricalDtype(categories=['a', 'b'], ordered=False)
NA and NaT group handling
If there are any NaN or NaT values in the grouping key, these will be automatically excluded. In other words, there will never be an “NA group” or “NaT group”. This was not the case in older versions of pandas, but users were generally discarding the NA group anyway (and supporting it was an implementation headache).
Grouping with ordered factors
Categorical variables represented as instance of pandas’s Categorical class
can be used as group keys. If so, the order of the levels will be preserved:
In [168]: data = pd.Series(np.random.randn(100))In [169]: factor = pd.qcut(data, [0, .25, .5, .75, 1.])In [170]: data.groupby(factor).mean()Out[170]:(-2.645, -0.523] -1.362896(-0.523, 0.0296] -0.260266(0.0296, 0.654] 0.361802(0.654, 2.21] 1.073801dtype: float64
Grouping with a grouper specification
You may need to specify a bit more data to properly group. You can
use the pd.Grouper to provide this local control.
In [171]: import datetimeIn [172]: df = pd.DataFrame({'Branch': 'A A A A A A A B'.split(),.....: 'Buyer': 'Carl Mark Carl Carl Joe Joe Joe Carl'.split(),.....: 'Quantity': [1, 3, 5, 1, 8, 1, 9, 3],.....: 'Date': [.....: datetime.datetime(2013, 1, 1, 13, 0),.....: datetime.datetime(2013, 1, 1, 13, 5),.....: datetime.datetime(2013, 10, 1, 20, 0),.....: datetime.datetime(2013, 10, 2, 10, 0),.....: datetime.datetime(2013, 10, 1, 20, 0),.....: datetime.datetime(2013, 10, 2, 10, 0),.....: datetime.datetime(2013, 12, 2, 12, 0),.....: datetime.datetime(2013, 12, 2, 14, 0)].....: }).....:In [173]: dfOut[173]:Branch Buyer Quantity Date0 A Carl 1 2013-01-01 13:00:001 A Mark 3 2013-01-01 13:05:002 A Carl 5 2013-10-01 20:00:003 A Carl 1 2013-10-02 10:00:004 A Joe 8 2013-10-01 20:00:005 A Joe 1 2013-10-02 10:00:006 A Joe 9 2013-12-02 12:00:007 B Carl 3 2013-12-02 14:00:00
Groupby a specific column with the desired frequency. This is like resampling.
In [174]: df.groupby([pd.Grouper(freq='1M', key='Date'), 'Buyer']).sum()Out[174]:QuantityDate Buyer2013-01-31 Carl 1Mark 32013-10-31 Carl 6Joe 92013-12-31 Carl 3Joe 9
You have an ambiguous specification in that you have a named index and a column that could be potential groupers.
In [175]: df = df.set_index('Date')In [176]: df['Date'] = df.index + pd.offsets.MonthEnd(2)In [177]: df.groupby([pd.Grouper(freq='6M', key='Date'), 'Buyer']).sum()Out[177]:QuantityDate Buyer2013-02-28 Carl 1Mark 32014-02-28 Carl 9Joe 18In [178]: df.groupby([pd.Grouper(freq='6M', level='Date'), 'Buyer']).sum()Out[178]:QuantityDate Buyer2013-01-31 Carl 1Mark 32014-01-31 Carl 9Joe 18
Taking the first rows of each group
Just like for a DataFrame or Series you can call head and tail on a groupby:
In [179]: df = pd.DataFrame([[1, 2], [1, 4], [5, 6]], columns=['A', 'B'])In [180]: dfOut[180]:A B0 1 21 1 42 5 6In [181]: g = df.groupby('A')In [182]: g.head(1)Out[182]:A B0 1 22 5 6In [183]: g.tail(1)Out[183]:A B1 1 42 5 6
This shows the first or last n rows from each group.
Taking the nth row of each group
To select from a DataFrame or Series the nth item, use
nth(). This is a reduction method, and
will return a single row (or no row) per group if you pass an int for n:
In [184]: df = pd.DataFrame([[1, np.nan], [1, 4], [5, 6]], columns=['A', 'B'])In [185]: g = df.groupby('A')In [186]: g.nth(0)Out[186]:BA1 NaN5 6.0In [187]: g.nth(-1)Out[187]:BA1 4.05 6.0In [188]: g.nth(1)Out[188]:BA1 4.0
If you want to select the nth not-null item, use the dropna kwarg. For a DataFrame this should be either 'any' or 'all' just like you would pass to dropna:
# nth(0) is the same as g.first()In [189]: g.nth(0, dropna='any')Out[189]:BA1 4.05 6.0In [190]: g.first()Out[190]:BA1 4.05 6.0# nth(-1) is the same as g.last()In [191]: g.nth(-1, dropna='any') # NaNs denote group exhausted when using dropnaOut[191]:BA1 4.05 6.0In [192]: g.last()Out[192]:BA1 4.05 6.0In [193]: g.B.nth(0, dropna='all')Out[193]:A1 4.05 6.0Name: B, dtype: float64
As with other methods, passing as_index=False, will achieve a filtration, which returns the grouped row.
In [194]: df = pd.DataFrame([[1, np.nan], [1, 4], [5, 6]], columns=['A', 'B'])In [195]: g = df.groupby('A', as_index=False)In [196]: g.nth(0)Out[196]:A B0 1 NaN2 5 6.0In [197]: g.nth(-1)Out[197]:A B1 1 4.02 5 6.0
You can also select multiple rows from each group by specifying multiple nth values as a list of ints.
In [198]: business_dates = pd.date_range(start='4/1/2014', end='6/30/2014', freq='B')In [199]: df = pd.DataFrame(1, index=business_dates, columns=['a', 'b'])# get the first, 4th, and last date index for each monthIn [200]: df.groupby([df.index.year, df.index.month]).nth([0, 3, -1])Out[200]:a b2014 4 1 14 1 14 1 15 1 15 1 15 1 16 1 16 1 16 1 1
Enumerate group items
To see the order in which each row appears within its group, use the
cumcount method:
In [201]: dfg = pd.DataFrame(list('aaabba'), columns=['A'])In [202]: dfgOut[202]:A0 a1 a2 a3 b4 b5 aIn [203]: dfg.groupby('A').cumcount()Out[203]:0 01 12 23 04 15 3dtype: int64In [204]: dfg.groupby('A').cumcount(ascending=False)Out[204]:0 31 22 13 14 05 0dtype: int64
Enumerate groups
New in version 0.20.2.
To see the ordering of the groups (as opposed to the order of rows
within a group given by cumcount) you can use
ngroup().
Note that the numbers given to the groups match the order in which the groups would be seen when iterating over the groupby object, not the order they are first observed.
In [205]: dfg = pd.DataFrame(list('aaabba'), columns=['A'])In [206]: dfgOut[206]:A0 a1 a2 a3 b4 b5 aIn [207]: dfg.groupby('A').ngroup()Out[207]:0 01 02 03 14 15 0dtype: int64In [208]: dfg.groupby('A').ngroup(ascending=False)Out[208]:0 11 12 13 04 05 1dtype: int64
Plotting
Groupby also works with some plotting methods. For example, suppose we suspect that some features in a DataFrame may differ by group, in this case, the values in column 1 where the group is “B” are 3 higher on average.
In [209]: np.random.seed(1234)In [210]: df = pd.DataFrame(np.random.randn(50, 2))In [211]: df['g'] = np.random.choice(['A', 'B'], size=50)In [212]: df.loc[df['g'] == 'B', 1] += 3
We can easily visualize this with a boxplot:
In [213]: df.groupby('g').boxplot()Out[213]:A AxesSubplot(0.1,0.15;0.363636x0.75)B AxesSubplot(0.536364,0.15;0.363636x0.75)dtype: object
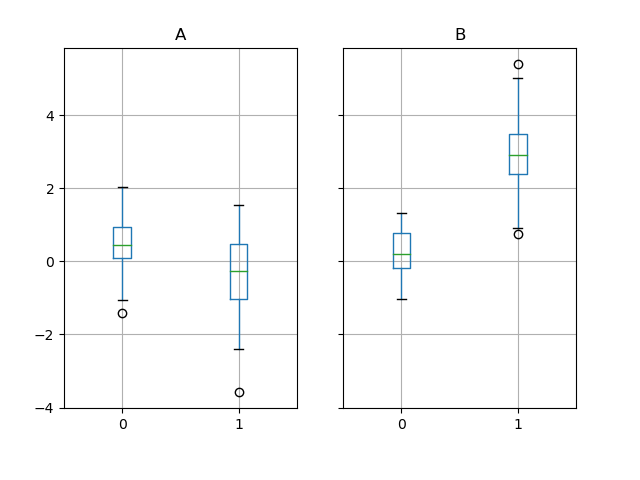
The result of calling boxplot is a dictionary whose keys are the values
of our grouping column g (“A” and “B”). The values of the resulting dictionary
can be controlled by the return_type keyword of boxplot.
See the visualization documentation for more.
::: danger Warning
For historical reasons, df.groupby("g").boxplot() is not equivalent
to df.boxplot(by="g"). See here for
an explanation.
:::
Piping function calls
New in version 0.21.0.
Similar to the functionality provided by DataFrame and Series, functions
that take GroupBy objects can be chained together using a pipe method to
allow for a cleaner, more readable syntax. To read about .pipe in general terms,
see here.
Combining .groupby and .pipe is often useful when you need to reuse
GroupBy objects.
As an example, imagine having a DataFrame with columns for stores, products, revenue and quantity sold. We’d like to do a groupwise calculation of prices (i.e. revenue/quantity) per store and per product. We could do this in a multi-step operation, but expressing it in terms of piping can make the code more readable. First we set the data:
In [214]: n = 1000In [215]: df = pd.DataFrame({'Store': np.random.choice(['Store_1', 'Store_2'], n),.....: 'Product': np.random.choice(['Product_1',.....: 'Product_2'], n),.....: 'Revenue': (np.random.random(n) * 50 + 10).round(2),.....: 'Quantity': np.random.randint(1, 10, size=n)}).....:In [216]: df.head(2)Out[216]:Store Product Revenue Quantity0 Store_2 Product_1 26.12 11 Store_2 Product_1 28.86 1
Now, to find prices per store/product, we can simply do:
In [217]: (df.groupby(['Store', 'Product']).....: .pipe(lambda grp: grp.Revenue.sum() / grp.Quantity.sum()).....: .unstack().round(2)).....:Out[217]:Product Product_1 Product_2StoreStore_1 6.82 7.05Store_2 6.30 6.64
Piping can also be expressive when you want to deliver a grouped object to some arbitrary function, for example:
In [218]: def mean(groupby):.....: return groupby.mean().....:In [219]: df.groupby(['Store', 'Product']).pipe(mean)Out[219]:Revenue QuantityStore ProductStore_1 Product_1 34.622727 5.075758Product_2 35.482815 5.029630Store_2 Product_1 32.972837 5.237589Product_2 34.684360 5.224000
where mean takes a GroupBy object and finds the mean of the Revenue and Quantity
columns respectively for each Store-Product combination. The mean function can
be any function that takes in a GroupBy object; the .pipe will pass the GroupBy
object as a parameter into the function you specify.
Examples
Regrouping by factor
Regroup columns of a DataFrame according to their sum, and sum the aggregated ones.
In [220]: df = pd.DataFrame({'a': [1, 0, 0], 'b': [0, 1, 0],.....: 'c': [1, 0, 0], 'd': [2, 3, 4]}).....:In [221]: dfOut[221]:a b c d0 1 0 1 21 0 1 0 32 0 0 0 4In [222]: df.groupby(df.sum(), axis=1).sum()Out[222]:1 90 2 21 1 32 0 4
Multi-column factorization
By using ngroup(), we can extract
information about the groups in a way similar to factorize() (as described
further in the reshaping API) but which applies
naturally to multiple columns of mixed type and different
sources. This can be useful as an intermediate categorical-like step
in processing, when the relationships between the group rows are more
important than their content, or as input to an algorithm which only
accepts the integer encoding. (For more information about support in
pandas for full categorical data, see the Categorical
introduction and the
API documentation.)
In [223]: dfg = pd.DataFrame({"A": [1, 1, 2, 3, 2], "B": list("aaaba")})In [224]: dfgOut[224]:A B0 1 a1 1 a2 2 a3 3 b4 2 aIn [225]: dfg.groupby(["A", "B"]).ngroup()Out[225]:0 01 02 13 24 1dtype: int64In [226]: dfg.groupby(["A", [0, 0, 0, 1, 1]]).ngroup()Out[226]:0 01 02 13 34 2dtype: int64
Groupby by indexer to ‘resample’ data
Resampling produces new hypothetical samples (resamples) from already existing observed data or from a model that generates data. These new samples are similar to the pre-existing samples.
In order to resample to work on indices that are non-datetimelike, the following procedure can be utilized.
In the following examples, df.index // 5 returns a binary array which is used to determine what gets selected for the groupby operation.
::: tip Note
The below example shows how we can downsample by consolidation of samples into fewer samples. Here by using df.index // 5, we are aggregating the samples in bins. By applying std() function, we aggregate the information contained in many samples into a small subset of values which is their standard deviation thereby reducing the number of samples.
:::
In [227]: df = pd.DataFrame(np.random.randn(10, 2))In [228]: dfOut[228]:0 10 -0.793893 0.3211531 0.342250 1.6189062 -0.975807 1.9182013 -0.810847 -1.4059194 -1.977759 0.4616595 0.730057 -1.3169386 -0.751328 0.5282907 -0.257759 -1.0810098 0.505895 -1.7019489 -1.006349 0.020208In [229]: df.index // 5Out[229]: Int64Index([0, 0, 0, 0, 0, 1, 1, 1, 1, 1], dtype='int64')In [230]: df.groupby(df.index // 5).std()Out[230]:0 10 0.823647 1.3129121 0.760109 0.942941
Returning a Series to propagate names
Group DataFrame columns, compute a set of metrics and return a named Series. The Series name is used as the name for the column index. This is especially useful in conjunction with reshaping operations such as stacking in which the column index name will be used as the name of the inserted column:
In [231]: df = pd.DataFrame({'a': [0, 0, 0, 0, 1, 1, 1, 1, 2, 2, 2, 2],.....: 'b': [0, 0, 1, 1, 0, 0, 1, 1, 0, 0, 1, 1],.....: 'c': [1, 0, 1, 0, 1, 0, 1, 0, 1, 0, 1, 0],.....: 'd': [0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 1]}).....:In [232]: def compute_metrics(x):.....: result = {'b_sum': x['b'].sum(), 'c_mean': x['c'].mean()}.....: return pd.Series(result, name='metrics').....:In [233]: result = df.groupby('a').apply(compute_metrics)In [234]: resultOut[234]:metrics b_sum c_meana0 2.0 0.51 2.0 0.52 2.0 0.5In [235]: result.stack()Out[235]:a metrics0 b_sum 2.0c_mean 0.51 b_sum 2.0c_mean 0.52 b_sum 2.0c_mean 0.5dtype: float64

