Computational tools
Statistical functions
Percent change
Series and DataFrame have a method
pct_change() to compute the percent change over a given number
of periods (using fill_method to fill NA/null values before computing
the percent change).
In [1]: ser = pd.Series(np.random.randn(8))In [2]: ser.pct_change()Out[2]:0 NaN1 -1.6029762 4.3349383 -0.2474564 -2.0673455 -1.1429036 -1.6882147 -9.759729dtype: float64
In [3]: df = pd.DataFrame(np.random.randn(10, 4))In [4]: df.pct_change(periods=3)Out[4]:0 1 2 30 NaN NaN NaN NaN1 NaN NaN NaN NaN2 NaN NaN NaN NaN3 -0.218320 -1.054001 1.987147 -0.5101834 -0.439121 -1.816454 0.649715 -4.8228095 -0.127833 -3.042065 -5.866604 -1.7769776 -2.596833 -1.959538 -2.111697 -3.7989007 -0.117826 -2.169058 0.036094 -0.0676968 2.492606 -1.357320 -1.205802 -1.5586979 -1.012977 2.324558 -1.003744 -0.371806
Covariance
Series.cov() can be used to compute covariance between series
(excluding missing values).
In [5]: s1 = pd.Series(np.random.randn(1000))In [6]: s2 = pd.Series(np.random.randn(1000))In [7]: s1.cov(s2)Out[7]: 0.000680108817431082
Analogously, DataFrame.cov() to compute pairwise covariances among the
series in the DataFrame, also excluding NA/null values.
::: tip Note
Assuming the missing data are missing at random this results in an estimate for the covariance matrix which is unbiased. However, for many applications this estimate may not be acceptable because the estimated covariance matrix is not guaranteed to be positive semi-definite. This could lead to estimated correlations having absolute values which are greater than one, and/or a non-invertible covariance matrix. See Estimation of covariance matrices for more details.
:::
In [8]: frame = pd.DataFrame(np.random.randn(1000, 5),...: columns=['a', 'b', 'c', 'd', 'e'])...:In [9]: frame.cov()Out[9]:a b c d ea 1.000882 -0.003177 -0.002698 -0.006889 0.031912b -0.003177 1.024721 0.000191 0.009212 0.000857c -0.002698 0.000191 0.950735 -0.031743 -0.005087d -0.006889 0.009212 -0.031743 1.002983 -0.047952e 0.031912 0.000857 -0.005087 -0.047952 1.042487
DataFrame.cov also supports an optional min_periods keyword that
specifies the required minimum number of observations for each column pair
in order to have a valid result.
In [10]: frame = pd.DataFrame(np.random.randn(20, 3), columns=['a', 'b', 'c'])In [11]: frame.loc[frame.index[:5], 'a'] = np.nanIn [12]: frame.loc[frame.index[5:10], 'b'] = np.nanIn [13]: frame.cov()Out[13]:a b ca 1.123670 -0.412851 0.018169b -0.412851 1.154141 0.305260c 0.018169 0.305260 1.301149In [14]: frame.cov(min_periods=12)Out[14]:a b ca 1.123670 NaN 0.018169b NaN 1.154141 0.305260c 0.018169 0.305260 1.301149
Correlation
Correlation may be computed using the corr() method.
Using the method parameter, several methods for computing correlations are
provided:
| Method name | Description |
|---|---|
| pearson (default) | Standard correlation coefficient |
| kendall | Kendall Tau correlation coefficient |
| spearman | Spearman rank correlation coefficient |
All of these are currently computed using pairwise complete observations. Wikipedia has articles covering the above correlation coefficients:
- Pearson correlation coefficient
- Kendall rank correlation coefficient
- Spearman’s rank correlation coefficient
::: tip Note
Please see the caveats associated with this method of calculating correlation matrices in the covariance section.
:::
In [15]: frame = pd.DataFrame(np.random.randn(1000, 5),....: columns=['a', 'b', 'c', 'd', 'e'])....:In [16]: frame.iloc[::2] = np.nan# Series with SeriesIn [17]: frame['a'].corr(frame['b'])Out[17]: 0.013479040400098794In [18]: frame['a'].corr(frame['b'], method='spearman')Out[18]: -0.007289885159540637# Pairwise correlation of DataFrame columnsIn [19]: frame.corr()Out[19]:a b c d ea 1.000000 0.013479 -0.049269 -0.042239 -0.028525b 0.013479 1.000000 -0.020433 -0.011139 0.005654c -0.049269 -0.020433 1.000000 0.018587 -0.054269d -0.042239 -0.011139 0.018587 1.000000 -0.017060e -0.028525 0.005654 -0.054269 -0.017060 1.000000
Note that non-numeric columns will be automatically excluded from the correlation calculation.
Like cov, corr also supports the optional min_periods keyword:
In [20]: frame = pd.DataFrame(np.random.randn(20, 3), columns=['a', 'b', 'c'])In [21]: frame.loc[frame.index[:5], 'a'] = np.nanIn [22]: frame.loc[frame.index[5:10], 'b'] = np.nanIn [23]: frame.corr()Out[23]:a b ca 1.000000 -0.121111 0.069544b -0.121111 1.000000 0.051742c 0.069544 0.051742 1.000000In [24]: frame.corr(min_periods=12)Out[24]:a b ca 1.000000 NaN 0.069544b NaN 1.000000 0.051742c 0.069544 0.051742 1.000000
New in version 0.24.0.
The method argument can also be a callable for a generic correlation
calculation. In this case, it should be a single function
that produces a single value from two ndarray inputs. Suppose we wanted to
compute the correlation based on histogram intersection:
# histogram intersectionIn [25]: def histogram_intersection(a, b):....: return np.minimum(np.true_divide(a, a.sum()),....: np.true_divide(b, b.sum())).sum()....:In [26]: frame.corr(method=histogram_intersection)Out[26]:a b ca 1.000000 -6.404882 -2.058431b -6.404882 1.000000 -19.255743c -2.058431 -19.255743 1.000000
A related method corrwith() is implemented on DataFrame to
compute the correlation between like-labeled Series contained in different
DataFrame objects.
In [27]: index = ['a', 'b', 'c', 'd', 'e']In [28]: columns = ['one', 'two', 'three', 'four']In [29]: df1 = pd.DataFrame(np.random.randn(5, 4), index=index, columns=columns)In [30]: df2 = pd.DataFrame(np.random.randn(4, 4), index=index[:4], columns=columns)In [31]: df1.corrwith(df2)Out[31]:one -0.125501two -0.493244three 0.344056four 0.004183dtype: float64In [32]: df2.corrwith(df1, axis=1)Out[32]:a -0.675817b 0.458296c 0.190809d -0.186275e NaNdtype: float64
Data ranking
The rank() method produces a data ranking with ties being
assigned the mean of the ranks (by default) for the group:
In [33]: s = pd.Series(np.random.np.random.randn(5), index=list('abcde'))In [34]: s['d'] = s['b'] # so there's a tieIn [35]: s.rank()Out[35]:a 5.0b 2.5c 1.0d 2.5e 4.0dtype: float64
rank() is also a DataFrame method and can rank either the rows
(axis=0) or the columns (axis=1). NaN values are excluded from the
ranking.
In [36]: df = pd.DataFrame(np.random.np.random.randn(10, 6))In [37]: df[4] = df[2][:5] # some tiesIn [38]: dfOut[38]:0 1 2 3 4 50 -0.904948 -1.163537 -1.457187 0.135463 -1.457187 0.2946501 -0.976288 -0.244652 -0.748406 -0.999601 -0.748406 -0.8008092 0.401965 1.460840 1.256057 1.308127 1.256057 0.8760043 0.205954 0.369552 -0.669304 0.038378 -0.669304 1.1402964 -0.477586 -0.730705 -1.129149 -0.601463 -1.129149 -0.2111965 -1.092970 -0.689246 0.908114 0.204848 NaN 0.4633476 0.376892 0.959292 0.095572 -0.593740 NaN -0.0691807 -1.002601 1.957794 -0.120708 0.094214 NaN -1.4674228 -0.547231 0.664402 -0.519424 -0.073254 NaN -1.2635449 -0.250277 -0.237428 -1.056443 0.419477 NaN 1.375064In [39]: df.rank(1)Out[39]:0 1 2 3 4 50 4.0 3.0 1.5 5.0 1.5 6.01 2.0 6.0 4.5 1.0 4.5 3.02 1.0 6.0 3.5 5.0 3.5 2.03 4.0 5.0 1.5 3.0 1.5 6.04 5.0 3.0 1.5 4.0 1.5 6.05 1.0 2.0 5.0 3.0 NaN 4.06 4.0 5.0 3.0 1.0 NaN 2.07 2.0 5.0 3.0 4.0 NaN 1.08 2.0 5.0 3.0 4.0 NaN 1.09 2.0 3.0 1.0 4.0 NaN 5.0
rank optionally takes a parameter ascending which by default is true;
when false, data is reverse-ranked, with larger values assigned a smaller rank.
rank supports different tie-breaking methods, specified with the method
parameter:
average: average rank of tied groupmin: lowest rank in the groupmax: highest rank in the groupfirst: ranks assigned in the order they appear in the array
Window Functions
For working with data, a number of window functions are provided for computing common window or rolling statistics. Among these are count, sum, mean, median, correlation, variance, covariance, standard deviation, skewness, and kurtosis.
The rolling() and expanding()
functions can be used directly from DataFrameGroupBy objects,
see the groupby docs.
::: tip Note
The API for window statistics is quite similar to the way one works with GroupBy objects, see the documentation here.
:::
We work with rolling, expanding and exponentially weighted data through the corresponding
objects, Rolling, Expanding and EWM.
In [40]: s = pd.Series(np.random.randn(1000),....: index=pd.date_range('1/1/2000', periods=1000))....:In [41]: s = s.cumsum()In [42]: sOut[42]:2000-01-01 -0.2688242000-01-02 -1.7718552000-01-03 -0.8180032000-01-04 -0.6592442000-01-05 -1.942133...2002-09-22 -67.4573232002-09-23 -69.2531822002-09-24 -70.2968182002-09-25 -70.8446742002-09-26 -72.475016Freq: D, Length: 1000, dtype: float64
These are created from methods on Series and DataFrame.
In [43]: r = s.rolling(window=60)In [44]: rOut[44]: Rolling [window=60,center=False,axis=0]
These object provide tab-completion of the available methods and properties.
In [14]: r.<TAB> # noqa: E225, E999r.agg r.apply r.count r.exclusions r.max r.median r.name r.skew r.sumr.aggregate r.corr r.cov r.kurt r.mean r.min r.quantile r.std r.var
Generally these methods all have the same interface. They all accept the following arguments:
window: size of moving windowmin_periods: threshold of non-null data points to require (otherwise result is NA)center: boolean, whether to set the labels at the center (default is False)
We can then call methods on these rolling objects. These return like-indexed objects:
In [45]: r.mean()Out[45]:2000-01-01 NaN2000-01-02 NaN2000-01-03 NaN2000-01-04 NaN2000-01-05 NaN...2002-09-22 -62.9149712002-09-23 -63.0618672002-09-24 -63.2138762002-09-25 -63.3750742002-09-26 -63.539734Freq: D, Length: 1000, dtype: float64
In [46]: s.plot(style='k--')Out[46]: <matplotlib.axes._subplots.AxesSubplot at 0x7f66048bdef0>In [47]: r.mean().plot(style='k')Out[47]: <matplotlib.axes._subplots.AxesSubplot at 0x7f66048bdef0>
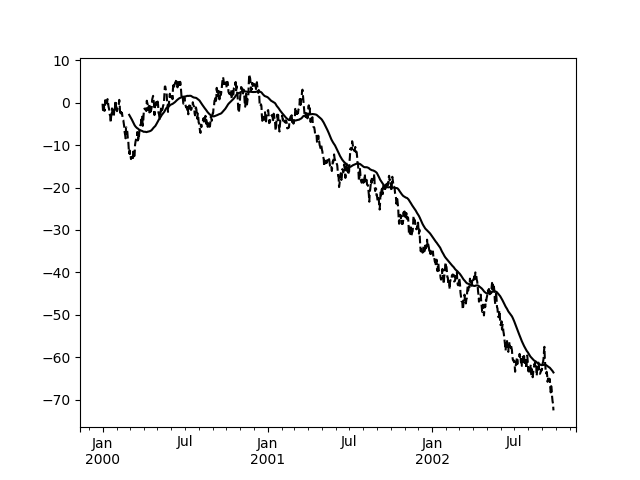
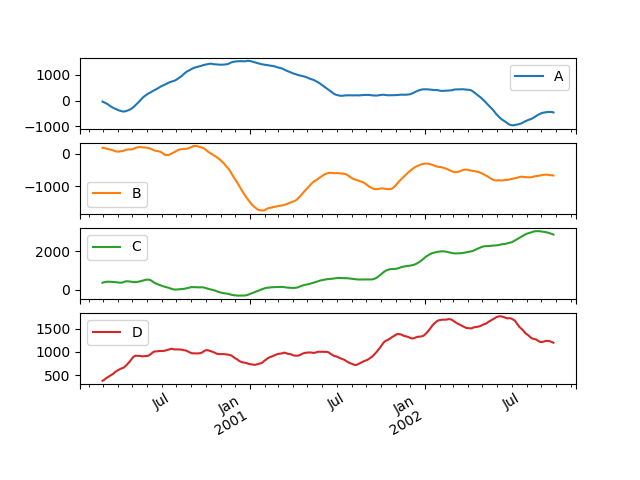
They can also be applied to DataFrame objects. This is really just syntactic sugar for applying the moving window operator to all of the DataFrame’s columns:
In [48]: df = pd.DataFrame(np.random.randn(1000, 4),....: index=pd.date_range('1/1/2000', periods=1000),....: columns=['A', 'B', 'C', 'D'])....:In [49]: df = df.cumsum()In [50]: df.rolling(window=60).sum().plot(subplots=True)Out[50]:array([<matplotlib.axes._subplots.AxesSubplot object at 0x7f66075a7f60>,<matplotlib.axes._subplots.AxesSubplot object at 0x7f65f79e55c0>,<matplotlib.axes._subplots.AxesSubplot object at 0x7f65f7998588>,<matplotlib.axes._subplots.AxesSubplot object at 0x7f65f794b550>],dtype=object)
Method summary
We provide a number of common statistical functions:
| Method | Description |
|---|---|
| count() | Number of non-null observations |
| sum() | Sum of values |
| mean() | Mean of values |
| median() | Arithmetic median of values |
| min() | Minimum |
| max() | Maximum |
| std() | Bessel-corrected sample standard deviation |
| var() | Unbiased variance |
| skew() | Sample skewness (3rd moment) |
| kurt() | Sample kurtosis (4th moment) |
| quantile() | Sample quantile (value at %) |
| apply() | Generic apply |
| cov() | Unbiased covariance (binary) |
| corr() | Correlation (binary) |
The apply() function takes an extra func argument and performs
generic rolling computations. The func argument should be a single function
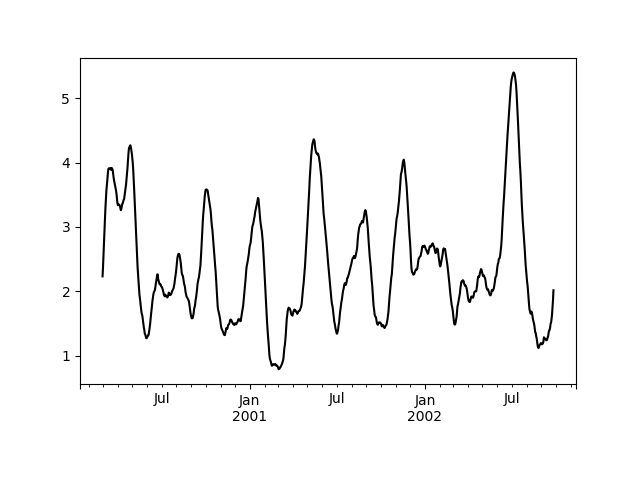
that produces a single value from an ndarray input. Suppose we wanted to
compute the mean absolute deviation on a rolling basis:
In [51]: def mad(x):....: return np.fabs(x - x.mean()).mean()....:In [52]: s.rolling(window=60).apply(mad, raw=True).plot(style='k')Out[52]: <matplotlib.axes._subplots.AxesSubplot at 0x7f65f795fac8>
Rolling windows
Passing win_type to .rolling generates a generic rolling window computation, that is weighted according the win_type.
The following methods are available:
| Method | Description |
|---|---|
| sum() | Sum of values |
| mean() | Mean of values |
The weights used in the window are specified by the win_type keyword.
The list of recognized types are the scipy.signal window functions:
boxcartriangblackmanhammingbartlettparzenbohmanblackmanharrisnuttallbarthannkaiser(needs beta)gaussian(needs std)general_gaussian(needs power, width)slepian(needs width)exponential(needs tau).
In [53]: ser = pd.Series(np.random.randn(10),....: index=pd.date_range('1/1/2000', periods=10))....:In [54]: ser.rolling(window=5, win_type='triang').mean()Out[54]:2000-01-01 NaN2000-01-02 NaN2000-01-03 NaN2000-01-04 NaN2000-01-05 -1.0378702000-01-06 -0.7677052000-01-07 -0.3831972000-01-08 -0.3955132000-01-09 -0.5584402000-01-10 -0.672416Freq: D, dtype: float64
Note that the boxcar window is equivalent to mean().
In [55]: ser.rolling(window=5, win_type='boxcar').mean()Out[55]:2000-01-01 NaN2000-01-02 NaN2000-01-03 NaN2000-01-04 NaN2000-01-05 -0.8411642000-01-06 -0.7799482000-01-07 -0.5654872000-01-08 -0.5028152000-01-09 -0.5537552000-01-10 -0.472211Freq: D, dtype: float64In [56]: ser.rolling(window=5).mean()Out[56]:2000-01-01 NaN2000-01-02 NaN2000-01-03 NaN2000-01-04 NaN2000-01-05 -0.8411642000-01-06 -0.7799482000-01-07 -0.5654872000-01-08 -0.5028152000-01-09 -0.5537552000-01-10 -0.472211Freq: D, dtype: float64
For some windowing functions, additional parameters must be specified:
In [57]: ser.rolling(window=5, win_type='gaussian').mean(std=0.1)Out[57]:2000-01-01 NaN2000-01-02 NaN2000-01-03 NaN2000-01-04 NaN2000-01-05 -1.3099892000-01-06 -1.1530002000-01-07 0.6063822000-01-08 -0.6811012000-01-09 -0.2897242000-01-10 -0.996632Freq: D, dtype: float64
::: tip Note
For .sum() with a win_type, there is no normalization done to the
weights for the window. Passing custom weights of [1, 1, 1] will yield a different
result than passing weights of [2, 2, 2], for example. When passing a
win_type instead of explicitly specifying the weights, the weights are
already normalized so that the largest weight is 1.
In contrast, the nature of the .mean() calculation is
such that the weights are normalized with respect to each other. Weights
of [1, 1, 1] and [2, 2, 2] yield the same result.
:::
Time-aware rolling
New in version 0.19.0.
New in version 0.19.0 are the ability to pass an offset (or convertible) to a .rolling() method and have it produce
variable sized windows based on the passed time window. For each time point, this includes all preceding values occurring
within the indicated time delta.
This can be particularly useful for a non-regular time frequency index.
In [58]: dft = pd.DataFrame({'B': [0, 1, 2, np.nan, 4]},....: index=pd.date_range('20130101 09:00:00',....: periods=5,....: freq='s'))....:In [59]: dftOut[59]:B2013-01-01 09:00:00 0.02013-01-01 09:00:01 1.02013-01-01 09:00:02 2.02013-01-01 09:00:03 NaN2013-01-01 09:00:04 4.0
This is a regular frequency index. Using an integer window parameter works to roll along the window frequency.
In [60]: dft.rolling(2).sum()Out[60]:B2013-01-01 09:00:00 NaN2013-01-01 09:00:01 1.02013-01-01 09:00:02 3.02013-01-01 09:00:03 NaN2013-01-01 09:00:04 NaNIn [61]: dft.rolling(2, min_periods=1).sum()Out[61]:B2013-01-01 09:00:00 0.02013-01-01 09:00:01 1.02013-01-01 09:00:02 3.02013-01-01 09:00:03 2.02013-01-01 09:00:04 4.0
Specifying an offset allows a more intuitive specification of the rolling frequency.
In [62]: dft.rolling('2s').sum()Out[62]:B2013-01-01 09:00:00 0.02013-01-01 09:00:01 1.02013-01-01 09:00:02 3.02013-01-01 09:00:03 2.02013-01-01 09:00:04 4.0
Using a non-regular, but still monotonic index, rolling with an integer window does not impart any special calculation.
In [63]: dft = pd.DataFrame({'B': [0, 1, 2, np.nan, 4]},....: index=pd.Index([pd.Timestamp('20130101 09:00:00'),....: pd.Timestamp('20130101 09:00:02'),....: pd.Timestamp('20130101 09:00:03'),....: pd.Timestamp('20130101 09:00:05'),....: pd.Timestamp('20130101 09:00:06')],....: name='foo'))....:In [64]: dftOut[64]:Bfoo2013-01-01 09:00:00 0.02013-01-01 09:00:02 1.02013-01-01 09:00:03 2.02013-01-01 09:00:05 NaN2013-01-01 09:00:06 4.0In [65]: dft.rolling(2).sum()Out[65]:Bfoo2013-01-01 09:00:00 NaN2013-01-01 09:00:02 1.02013-01-01 09:00:03 3.02013-01-01 09:00:05 NaN2013-01-01 09:00:06 NaN
Using the time-specification generates variable windows for this sparse data.
In [66]: dft.rolling('2s').sum()Out[66]:Bfoo2013-01-01 09:00:00 0.02013-01-01 09:00:02 1.02013-01-01 09:00:03 3.02013-01-01 09:00:05 NaN2013-01-01 09:00:06 4.0
Furthermore, we now allow an optional on parameter to specify a column (rather than the
default of the index) in a DataFrame.
In [67]: dft = dft.reset_index()In [68]: dftOut[68]:foo B0 2013-01-01 09:00:00 0.01 2013-01-01 09:00:02 1.02 2013-01-01 09:00:03 2.03 2013-01-01 09:00:05 NaN4 2013-01-01 09:00:06 4.0In [69]: dft.rolling('2s', on='foo').sum()Out[69]:foo B0 2013-01-01 09:00:00 0.01 2013-01-01 09:00:02 1.02 2013-01-01 09:00:03 3.03 2013-01-01 09:00:05 NaN4 2013-01-01 09:00:06 4.0
Rolling window endpoints
New in version 0.20.0.
The inclusion of the interval endpoints in rolling window calculations can be specified with the closed
parameter:
| closed | Description | Default for |
|---|---|---|
| right | close right endpoint | time-based windows |
| left | close left endpoint | |
| both | close both endpoints | fixed windows |
| neither | open endpoints |
For example, having the right endpoint open is useful in many problems that require that there is no contamination from present information back to past information. This allows the rolling window to compute statistics “up to that point in time”, but not including that point in time.
In [70]: df = pd.DataFrame({'x': 1},....: index=[pd.Timestamp('20130101 09:00:01'),....: pd.Timestamp('20130101 09:00:02'),....: pd.Timestamp('20130101 09:00:03'),....: pd.Timestamp('20130101 09:00:04'),....: pd.Timestamp('20130101 09:00:06')])....:In [71]: df["right"] = df.rolling('2s', closed='right').x.sum() # defaultIn [72]: df["both"] = df.rolling('2s', closed='both').x.sum()In [73]: df["left"] = df.rolling('2s', closed='left').x.sum()In [74]: df["neither"] = df.rolling('2s', closed='neither').x.sum()In [75]: dfOut[75]:x right both left neither2013-01-01 09:00:01 1 1.0 1.0 NaN NaN2013-01-01 09:00:02 1 2.0 2.0 1.0 1.02013-01-01 09:00:03 1 2.0 3.0 2.0 1.02013-01-01 09:00:04 1 2.0 3.0 2.0 1.02013-01-01 09:00:06 1 1.0 2.0 1.0 NaN
Currently, this feature is only implemented for time-based windows. For fixed windows, the closed parameter cannot be set and the rolling window will always have both endpoints closed.
Time-aware rolling vs. resampling
Using .rolling() with a time-based index is quite similar to resampling. They
both operate and perform reductive operations on time-indexed pandas objects.
When using .rolling() with an offset. The offset is a time-delta. Take a backwards-in-time looking window, and
aggregate all of the values in that window (including the end-point, but not the start-point). This is the new value
at that point in the result. These are variable sized windows in time-space for each point of the input. You will get
a same sized result as the input.
When using .resample() with an offset. Construct a new index that is the frequency of the offset. For each frequency
bin, aggregate points from the input within a backwards-in-time looking window that fall in that bin. The result of this
aggregation is the output for that frequency point. The windows are fixed size in the frequency space. Your result
will have the shape of a regular frequency between the min and the max of the original input object.
To summarize, .rolling() is a time-based window operation, while .resample() is a frequency-based window operation.
Centering windows
By default the labels are set to the right edge of the window, but a
center keyword is available so the labels can be set at the center.
In [76]: ser.rolling(window=5).mean()Out[76]:2000-01-01 NaN2000-01-02 NaN2000-01-03 NaN2000-01-04 NaN2000-01-05 -0.8411642000-01-06 -0.7799482000-01-07 -0.5654872000-01-08 -0.5028152000-01-09 -0.5537552000-01-10 -0.472211Freq: D, dtype: float64In [77]: ser.rolling(window=5, center=True).mean()Out[77]:2000-01-01 NaN2000-01-02 NaN2000-01-03 -0.8411642000-01-04 -0.7799482000-01-05 -0.5654872000-01-06 -0.5028152000-01-07 -0.5537552000-01-08 -0.4722112000-01-09 NaN2000-01-10 NaNFreq: D, dtype: float64
Binary window functions
cov() and corr() can compute moving window statistics about
two Series or any combination of DataFrame/Series or
DataFrame/DataFrame. Here is the behavior in each case:
- two
Series: compute the statistic for the pairing. DataFrame/Series: compute the statistics for each column of the DataFrame with the passed Series, thus returning a DataFrame.DataFrame/DataFrame: by default compute the statistic for matching column names, returning a DataFrame. If the keyword argumentpairwise=Trueis passed then computes the statistic for each pair of columns, returning aMultiIndexed DataFramewhoseindexare the dates in question (see the next section).
For example:
In [78]: df = pd.DataFrame(np.random.randn(1000, 4),....: index=pd.date_range('1/1/2000', periods=1000),....: columns=['A', 'B', 'C', 'D'])....:In [79]: df = df.cumsum()In [80]: df2 = df[:20]In [81]: df2.rolling(window=5).corr(df2['B'])Out[81]:A B C D2000-01-01 NaN NaN NaN NaN2000-01-02 NaN NaN NaN NaN2000-01-03 NaN NaN NaN NaN2000-01-04 NaN NaN NaN NaN2000-01-05 0.768775 1.0 -0.977990 0.800252... ... ... ... ...2000-01-16 0.691078 1.0 0.807450 -0.9393022000-01-17 0.274506 1.0 0.582601 -0.9029542000-01-18 0.330459 1.0 0.515707 -0.5452682000-01-19 0.046756 1.0 -0.104334 -0.4197992000-01-20 -0.328241 1.0 -0.650974 -0.777777[20 rows x 4 columns]
Computing rolling pairwise covariances and correlations
In financial data analysis and other fields it’s common to compute covariance
and correlation matrices for a collection of time series. Often one is also
interested in moving-window covariance and correlation matrices. This can be
done by passing the pairwise keyword argument, which in the case of
DataFrame inputs will yield a MultiIndexed DataFrame whose index are the dates in
question. In the case of a single DataFrame argument the pairwise argument
can even be omitted:
::: tip Note
Missing values are ignored and each entry is computed using the pairwise complete observations. Please see the covariance section for caveats associated with this method of calculating covariance and correlation matrices.
:::
In [82]: covs = (df[['B', 'C', 'D']].rolling(window=50)....: .cov(df[['A', 'B', 'C']], pairwise=True))....:In [83]: covs.loc['2002-09-22':]Out[83]:B C D2002-09-22 A 1.367467 8.676734 -8.047366B 3.067315 0.865946 -1.052533C 0.865946 7.739761 -4.9439242002-09-23 A 0.910343 8.669065 -8.443062B 2.625456 0.565152 -0.907654C 0.565152 7.825521 -5.3675262002-09-24 A 0.463332 8.514509 -8.776514B 2.306695 0.267746 -0.732186C 0.267746 7.771425 -5.6969622002-09-25 A 0.467976 8.198236 -9.162599B 2.307129 0.267287 -0.754080C 0.267287 7.466559 -5.8226502002-09-26 A 0.545781 7.899084 -9.326238B 2.311058 0.322295 -0.844451C 0.322295 7.038237 -5.684445
In [84]: correls = df.rolling(window=50).corr()In [85]: correls.loc['2002-09-22':]Out[85]:A B C D2002-09-22 A 1.000000 0.186397 0.744551 -0.769767B 0.186397 1.000000 0.177725 -0.240802C 0.744551 0.177725 1.000000 -0.712051D -0.769767 -0.240802 -0.712051 1.0000002002-09-23 A 1.000000 0.134723 0.743113 -0.758758... ... ... ... ...2002-09-25 D -0.739160 -0.164179 -0.704686 1.0000002002-09-26 A 1.000000 0.087756 0.727792 -0.736562B 0.087756 1.000000 0.079913 -0.179477C 0.727792 0.079913 1.000000 -0.692303D -0.736562 -0.179477 -0.692303 1.000000[20 rows x 4 columns]
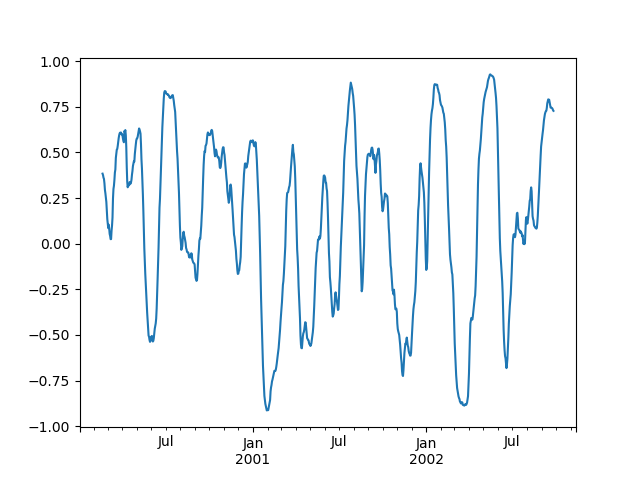
You can efficiently retrieve the time series of correlations between two columns by reshaping and indexing:
In [86]: correls.unstack(1)[('A', 'C')].plot()Out[86]: <matplotlib.axes._subplots.AxesSubplot at 0x7f65f8dc79e8>
Aggregation
Once the Rolling, Expanding or EWM objects have been created, several methods are available to
perform multiple computations on the data. These operations are similar to the aggregating API,
groupby API, and resample API.
In [87]: dfa = pd.DataFrame(np.random.randn(1000, 3),....: index=pd.date_range('1/1/2000', periods=1000),....: columns=['A', 'B', 'C'])....:In [88]: r = dfa.rolling(window=60, min_periods=1)In [89]: rOut[89]: Rolling [window=60,min_periods=1,center=False,axis=0]
We can aggregate by passing a function to the entire DataFrame, or select a
Series (or multiple Series) via standard __getitem__.
In [90]: r.aggregate(np.sum)Out[90]:A B C2000-01-01 -0.289838 -0.370545 -1.2842062000-01-02 -0.216612 -1.675528 -1.1694152000-01-03 1.154661 -1.634017 -1.5666202000-01-04 2.969393 -4.003274 -1.8161792000-01-05 4.690630 -4.682017 -2.717209... ... ... ...2002-09-22 2.860036 -9.270337 6.4152452002-09-23 3.510163 -8.151439 5.1772192002-09-24 6.524983 -10.168078 5.7926392002-09-25 6.409626 -9.956226 5.7040502002-09-26 5.093787 -7.074515 6.905823[1000 rows x 3 columns]In [91]: r['A'].aggregate(np.sum)Out[91]:2000-01-01 -0.2898382000-01-02 -0.2166122000-01-03 1.1546612000-01-04 2.9693932000-01-05 4.690630...2002-09-22 2.8600362002-09-23 3.5101632002-09-24 6.5249832002-09-25 6.4096262002-09-26 5.093787Freq: D, Name: A, Length: 1000, dtype: float64In [92]: r[['A', 'B']].aggregate(np.sum)Out[92]:A B2000-01-01 -0.289838 -0.3705452000-01-02 -0.216612 -1.6755282000-01-03 1.154661 -1.6340172000-01-04 2.969393 -4.0032742000-01-05 4.690630 -4.682017... ... ...2002-09-22 2.860036 -9.2703372002-09-23 3.510163 -8.1514392002-09-24 6.524983 -10.1680782002-09-25 6.409626 -9.9562262002-09-26 5.093787 -7.074515[1000 rows x 2 columns]
As you can see, the result of the aggregation will have the selected columns, or all columns if none are selected.
Applying multiple functions
With windowed Series you can also pass a list of functions to do
aggregation with, outputting a DataFrame:
In [93]: r['A'].agg([np.sum, np.mean, np.std])Out[93]:sum mean std2000-01-01 -0.289838 -0.289838 NaN2000-01-02 -0.216612 -0.108306 0.2567252000-01-03 1.154661 0.384887 0.8733112000-01-04 2.969393 0.742348 1.0097342000-01-05 4.690630 0.938126 0.977914... ... ... ...2002-09-22 2.860036 0.047667 1.1320512002-09-23 3.510163 0.058503 1.1342962002-09-24 6.524983 0.108750 1.1442042002-09-25 6.409626 0.106827 1.1429132002-09-26 5.093787 0.084896 1.151416[1000 rows x 3 columns]
On a windowed DataFrame, you can pass a list of functions to apply to each column, which produces an aggregated result with a hierarchical index:
In [94]: r.agg([np.sum, np.mean])Out[94]:A B Csum mean sum mean sum mean2000-01-01 -0.289838 -0.289838 -0.370545 -0.370545 -1.284206 -1.2842062000-01-02 -0.216612 -0.108306 -1.675528 -0.837764 -1.169415 -0.5847082000-01-03 1.154661 0.384887 -1.634017 -0.544672 -1.566620 -0.5222072000-01-04 2.969393 0.742348 -4.003274 -1.000819 -1.816179 -0.4540452000-01-05 4.690630 0.938126 -4.682017 -0.936403 -2.717209 -0.543442... ... ... ... ... ... ...2002-09-22 2.860036 0.047667 -9.270337 -0.154506 6.415245 0.1069212002-09-23 3.510163 0.058503 -8.151439 -0.135857 5.177219 0.0862872002-09-24 6.524983 0.108750 -10.168078 -0.169468 5.792639 0.0965442002-09-25 6.409626 0.106827 -9.956226 -0.165937 5.704050 0.0950682002-09-26 5.093787 0.084896 -7.074515 -0.117909 6.905823 0.115097[1000 rows x 6 columns]
Passing a dict of functions has different behavior by default, see the next section.
Applying different functions to DataFrame columns
By passing a dict to aggregate you can apply a different aggregation to the
columns of a DataFrame:
In [95]: r.agg({'A': np.sum, 'B': lambda x: np.std(x, ddof=1)})Out[95]:A B2000-01-01 -0.289838 NaN2000-01-02 -0.216612 0.6607472000-01-03 1.154661 0.6899292000-01-04 2.969393 1.0721992000-01-05 4.690630 0.939657... ... ...2002-09-22 2.860036 1.1132082002-09-23 3.510163 1.1323812002-09-24 6.524983 1.0809632002-09-25 6.409626 1.0829112002-09-26 5.093787 1.136199[1000 rows x 2 columns]
The function names can also be strings. In order for a string to be valid it must be implemented on the windowed object
In [96]: r.agg({'A': 'sum', 'B': 'std'})Out[96]:A B2000-01-01 -0.289838 NaN2000-01-02 -0.216612 0.6607472000-01-03 1.154661 0.6899292000-01-04 2.969393 1.0721992000-01-05 4.690630 0.939657... ... ...2002-09-22 2.860036 1.1132082002-09-23 3.510163 1.1323812002-09-24 6.524983 1.0809632002-09-25 6.409626 1.0829112002-09-26 5.093787 1.136199[1000 rows x 2 columns]
Furthermore you can pass a nested dict to indicate different aggregations on different columns.
In [97]: r.agg({'A': ['sum', 'std'], 'B': ['mean', 'std']})Out[97]:A Bsum std mean std2000-01-01 -0.289838 NaN -0.370545 NaN2000-01-02 -0.216612 0.256725 -0.837764 0.6607472000-01-03 1.154661 0.873311 -0.544672 0.6899292000-01-04 2.969393 1.009734 -1.000819 1.0721992000-01-05 4.690630 0.977914 -0.936403 0.939657... ... ... ... ...2002-09-22 2.860036 1.132051 -0.154506 1.1132082002-09-23 3.510163 1.134296 -0.135857 1.1323812002-09-24 6.524983 1.144204 -0.169468 1.0809632002-09-25 6.409626 1.142913 -0.165937 1.0829112002-09-26 5.093787 1.151416 -0.117909 1.136199[1000 rows x 4 columns]
Expanding windows
A common alternative to rolling statistics is to use an expanding window, which yields the value of the statistic with all the data available up to that point in time.
These follow a similar interface to .rolling, with the .expanding method
returning an Expanding object.
As these calculations are a special case of rolling statistics, they are implemented in pandas such that the following two calls are equivalent:
In [98]: df.rolling(window=len(df), min_periods=1).mean()[:5]Out[98]:A B C D2000-01-01 0.314226 -0.001675 0.071823 0.8925662000-01-02 0.654522 -0.171495 0.179278 0.8533612000-01-03 0.708733 -0.064489 -0.238271 1.3711112000-01-04 0.987613 0.163472 -0.919693 1.5664852000-01-05 1.426971 0.288267 -1.358877 1.808650In [99]: df.expanding(min_periods=1).mean()[:5]Out[99]:A B C D2000-01-01 0.314226 -0.001675 0.071823 0.8925662000-01-02 0.654522 -0.171495 0.179278 0.8533612000-01-03 0.708733 -0.064489 -0.238271 1.3711112000-01-04 0.987613 0.163472 -0.919693 1.5664852000-01-05 1.426971 0.288267 -1.358877 1.808650
These have a similar set of methods to .rolling methods.
Method summary
| Function | Description |
|---|---|
| count() | Number of non-null observations |
| sum() | Sum of values |
| mean() | Mean of values |
| median() | Arithmetic median of values |
| min() | Minimum |
| max() | Maximum |
| std() | Unbiased standard deviation |
| var() | Unbiased variance |
| skew() | Unbiased skewness (3rd moment) |
| kurt() | Unbiased kurtosis (4th moment) |
| quantile() | Sample quantile (value at %) |
| apply() | Generic apply |
| cov() | Unbiased covariance (binary) |
| corr() | Correlation (binary) |
Aside from not having a window parameter, these functions have the same
interfaces as their .rolling counterparts. Like above, the parameters they
all accept are:
min_periods: threshold of non-null data points to require. Defaults to minimum needed to compute statistic. NoNaNswill be output oncemin_periodsnon-null data points have been seen.center: boolean, whether to set the labels at the center (default is False).
::: tip Note
The output of the .rolling and .expanding methods do not return a
NaN if there are at least min_periods non-null values in the current
window. For example:
In [100]: sn = pd.Series([1, 2, np.nan, 3, np.nan, 4])In [101]: snOut[101]:0 1.01 2.02 NaN3 3.04 NaN5 4.0dtype: float64In [102]: sn.rolling(2).max()Out[102]:0 NaN1 2.02 NaN3 NaN4 NaN5 NaNdtype: float64In [103]: sn.rolling(2, min_periods=1).max()Out[103]:0 1.01 2.02 2.03 3.04 3.05 4.0dtype: float64
In case of expanding functions, this differs from cumsum(),
cumprod(), cummax(),
and cummin(), which return NaN in the output wherever
a NaN is encountered in the input. In order to match the output of cumsum
with expanding, use fillna():
In [104]: sn.expanding().sum()Out[104]:0 1.01 3.02 3.03 6.04 6.05 10.0dtype: float64In [105]: sn.cumsum()Out[105]:0 1.01 3.02 NaN3 6.04 NaN5 10.0dtype: float64In [106]: sn.cumsum().fillna(method='ffill')Out[106]:0 1.01 3.02 3.03 6.04 6.05 10.0dtype: float64
:::
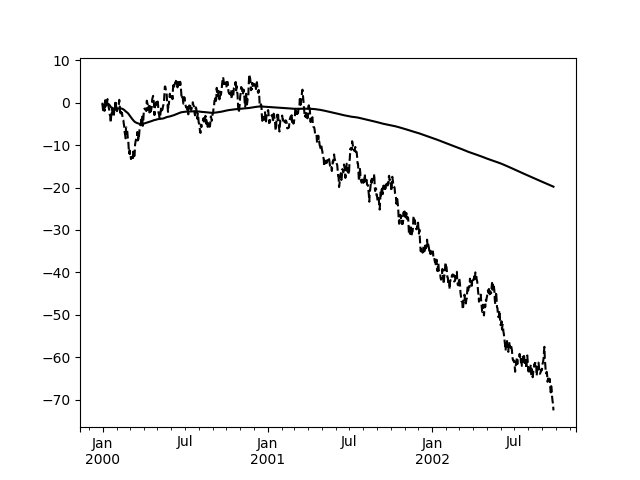
An expanding window statistic will be more stable (and less responsive) than
its rolling window counterpart as the increasing window size decreases the
relative impact of an individual data point. As an example, here is the
mean() output for the previous time series dataset:
In [107]: s.plot(style='k--')Out[107]: <matplotlib.axes._subplots.AxesSubplot at 0x7f6607b14400>In [108]: s.expanding().mean().plot(style='k')Out[108]: <matplotlib.axes._subplots.AxesSubplot at 0x7f6607b14400>
Exponentially weighted windows
A related set of functions are exponentially weighted versions of several of
the above statistics. A similar interface to .rolling and .expanding is accessed
through the .ewm method to receive an EWM object.
A number of expanding EW (exponentially weighted)
methods are provided:
| Function | Description |
|---|---|
| mean() | EW moving average |
| var() | EW moving variance |
| std() | EW moving standard deviation |
| corr() | EW moving correlation |
| cov() | EW moving covariance |
In general, a weighted moving average is calculated as
where \(x_t\) is the input, \(y_t\) is the result and the \(w_i\) are the weights.
The EW functions support two variants of exponential weights.
The default, adjust=True, uses the weights \(w_i = (1 - \alpha)^i\)
which gives
When adjust=False is specified, moving averages are calculated as
which is equivalent to using weights
::: tip Note
These equations are sometimes written in terms of \(\alpha’ = 1 - \alpha\), e.g.
:::
The difference between the above two variants arises because we are
dealing with series which have finite history. Consider a series of infinite
history, with adjust=True:
Noting that the denominator is a geometric series with initial term equal to 1 and a ratio of \(1 - \alpha\) we have
which is the same expression as adjust=False above and therefore
shows the equivalence of the two variants for infinite series.
When adjust=False, we have \(y0 = x_0\) and
\(y_t = \alpha x_t + (1 - \alpha) y{t-1}\).
Therefore, there is an assumption that \(x_0\) is not an ordinary value
but rather an exponentially weighted moment of the infinite series up to that
point.
One must have \(0 < \alpha \leq 1\), and while since version 0.18.0 it has been possible to pass \(\alpha\) directly, it’s often easier to think about either the span, center of mass (com) or half-life of an EW moment:
One must specify precisely one of span, center of mass, half-life and alpha to the EW functions:
- Span corresponds to what is commonly called an “N-day EW moving average”.
- Center of mass has a more physical interpretation and can be thought of in terms of span: \(c = (s - 1) / 2\).
- Half-life is the period of time for the exponential weight to reduce to one half.
- Alpha specifies the smoothing factor directly.
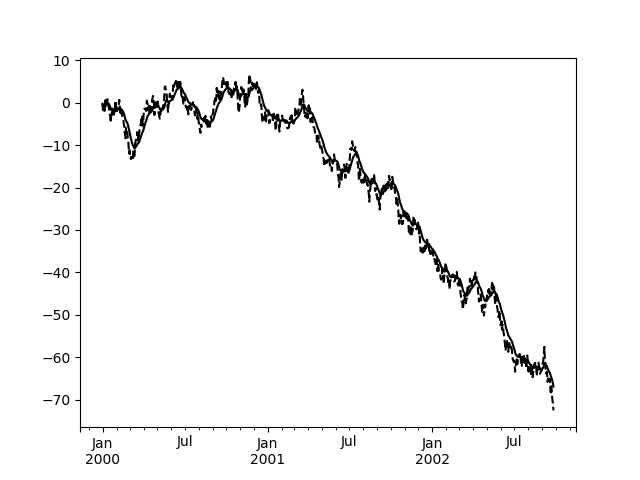
Here is an example for a univariate time series:
In [109]: s.plot(style='k--')Out[109]: <matplotlib.axes._subplots.AxesSubplot at 0x7f65f7537278>In [110]: s.ewm(span=20).mean().plot(style='k')Out[110]: <matplotlib.axes._subplots.AxesSubplot at 0x7f65f7537278>
EWM has a min_periods argument, which has the same
meaning it does for all the .expanding and .rolling methods:
no output values will be set until at least min_periods non-null values
are encountered in the (expanding) window.
EWM also has an ignore_na argument, which determines how
intermediate null values affect the calculation of the weights.
When ignore_na=False (the default), weights are calculated based on absolute
positions, so that intermediate null values affect the result.
When ignore_na=True,
weights are calculated by ignoring intermediate null values.
For example, assuming adjust=True, if ignore_na=False, the weighted
average of 3, NaN, 5 would be calculated as
Whereas if ignore_na=True, the weighted average would be calculated as
The var(), std(), and cov() functions have a bias argument,
specifying whether the result should contain biased or unbiased statistics.
For example, if bias=True, ewmvar(x) is calculated as
ewmvar(x) = ewma(x**2) - ewma(x)**2;
whereas if bias=False (the default), the biased variance statistics
are scaled by debiasing factors
(For \(w_i = 1\), this reduces to the usual \(N / (N - 1)\) factor, with \(N = t + 1\).) See Weighted Sample Variance on Wikipedia for further details.

