Aligning Similar Substrings
Whereas global alignment (see “Global Alignment with Scoring Matrix”) can be helpful for comparing genetic strings of similar length that resemble each other, often we will be presented with strings that are mostly dissimilar except for some unknown region of the strings, which may represent a shared gene. To find such genes, we need to modify global alignment to instead search for shared motifs in the form of locally similar regions (recall “Finding a Shared Motif” and “Finding a Shared Spliced Motif”).
Using global alignment often fails to find shared motifs hidden in larger strings because (especially if the similar region is found on different ends of the string) aligning the strings causes gap penalties to rack up.
If we are only interested in comparing the regions of similarity, then we would like to have some way of disregarding the parts of the strings that don’t resemble each other. The way to do this is to produce alignment scores for all possible pairs of substrings.
Problem
A local alignment of two strings s and t is an alignment of substrings r and u of s and t, respectively. Let opt(r,u) denote the score of an optimal alignment of r and u with respect to some predetermined alignment score.
Given: Two protein strings s and t in FASTA format (each having length at most 1000 aa).
Return: A maximum alignment score along with substrings r and u of s and t, respectively, which produce this maximum alignment score (multiple solutions may exist, in which case you may output any one). Use:
- The PAM250 scoring matrix.
- Linear gap penalty equal to 5.
Sample Dataset
Rosalind_80
MEANLYPRTEINSTRING
>Rosalind_21
PLEASANTLYEINSTEINSample Output
23
LYPRTEINSTRIN
LYEINSTEIN
A local alignment of two strings is an alignment of a substring of one of the strings with a substring of the other. Local alignment is in contrast with a global alignment, in which every symbol of both strings is aligned; as a result, in local alignment, only those symbols belonging to the substrings that are aligned will contribute to the calculation of the alignment’s score. This serves a practical purpose when we wish to search large genetic strings for small regions of the highest similarity.
In the figure below, an optimal global alignment of two DNA strings is shown, followed by an optimal local alignment of the same two strings. The edit alignment score function is used in both cases; the local alignment of the two strings has a net score equal to 0 because the symbols outside the matched interval are not counted. This figure demonstrates the importance of local alignment in applications, in which we often search very long genetic strings for small, local areas of similarity (e.g., finding shared genes along an interval of two chromosomes).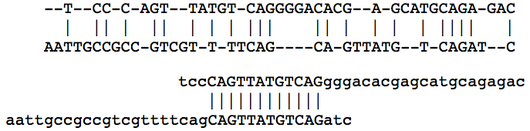
Solution
PAM250 is one of a family of alignment scoring matrices introduced by Margaret Dayhoff in 1978.
A C D E F G H I K L M N P Q R S T V W YA 2 -2 0 0 -3 1 -1 -1 -1 -2 -1 0 1 0 -2 1 1 0 -6 -3C -2 12 -5 -5 -4 -3 -3 -2 -5 -6 -5 -4 -3 -5 -4 0 -2 -2 -8 0D 0 -5 4 3 -6 1 1 -2 0 -4 -3 2 -1 2 -1 0 0 -2 -7 -4E 0 -5 3 4 -5 0 1 -2 0 -3 -2 1 -1 2 -1 0 0 -2 -7 -4F -3 -4 -6 -5 9 -5 -2 1 -5 2 0 -3 -5 -5 -4 -3 -3 -1 0 7G 1 -3 1 0 -5 5 -2 -3 -2 -4 -3 0 0 -1 -3 1 0 -1 -7 -5H -1 -3 1 1 -2 -2 6 -2 0 -2 -2 2 0 3 2 -1 -1 -2 -3 0I -1 -2 -2 -2 1 -3 -2 5 -2 2 2 -2 -2 -2 -2 -1 0 4 -5 -1K -1 -5 0 0 -5 -2 0 -2 5 -3 0 1 -1 1 3 0 0 -2 -3 -4L -2 -6 -4 -3 2 -4 -2 2 -3 6 4 -3 -3 -2 -3 -3 -2 2 -2 -1M -1 -5 -3 -2 0 -3 -2 2 0 4 6 -2 -2 -1 0 -2 -1 2 -4 -2N 0 -4 2 1 -3 0 2 -2 1 -3 -2 2 0 1 0 1 0 -2 -4 -2P 1 -3 -1 -1 -5 0 0 -2 -1 -3 -2 0 6 0 0 1 0 -1 -6 -5Q 0 -5 2 2 -5 -1 3 -2 1 -2 -1 1 0 4 1 -1 -1 -2 -5 -4R -2 -4 -1 -1 -4 -3 2 -2 3 -3 0 0 0 1 6 0 -1 -2 2 -4S 1 0 0 0 -3 1 -1 -1 0 -3 -2 1 1 -1 0 2 1 -1 -2 -3T 1 -2 0 0 -3 0 -1 0 0 -2 -1 0 0 -1 -1 1 3 0 -5 -3V 0 -2 -2 -2 -1 -1 -2 4 -2 2 2 -2 -1 -2 -2 -1 0 4 -6 -2W -6 -8 -7 -7 0 -7 -3 -5 -3 -2 -4 -4 -6 -5 2 -2 -5 -6 17 0Y -3 0 -4 -4 7 -5 0 -1 -4 -1 -2 -2 -5 -4 -4 -3 -3 -2 0 10
同全局比对算法一样,只不过将状态转移方程多增加一个 0 ,也就是局部高分不能因为插入空位终止。同时区别在于不再是从最右下角进行回溯,而是直接从 dp 表中最大的一个进行倒推。
from typing import Listchr2idx = lambda ch: ord(ch) - ord('A')class Solution:def localAlignment(self, s: str, t: str, table: List[List[int]]) -> List[str]:GAP = -5m, n = len(s), len(t)dp = [[0] * (n + 1) for _ in range(m + 1)]maxi = maxj = 0for i in range(1, m + 1):for j in range(1, n + 1):dp[i][j] = max(0,dp[i-1][j] + GAP,dp[i][j-1] + GAP,dp[i-1][j-1] + table[chr2idx(s[i-1])][chr2idx(t[j-1])])if dp[i][j] > dp[maxi][maxj]:maxi, maxj = i, jdef backtrace(i: int, j: int, path1: List[int], path2: List[int]) -> None:if i * j == 0 or dp[i][j] == 0:print(''.join(reversed(path1)))print(''.join(reversed(path2)))print()returnif dp[i][j] == dp[i-1][j] + GAP:path1.append(s[i-1])path2.append('-')backtrace(i - 1, j, path1, path2)path1.pop()path2.pop()if dp[i][j] == dp[i][j-1] + GAP:path1.append('-')path2.append(t[j-1])backtrace(i, j - 1, path1, path2)path1.pop()path2.pop()if dp[i][j] == dp[i-1][j-1] + table[chr2idx(s[i-1])][chr2idx(t[j-1])]:path1.append(s[i-1])path2.append(t[j-1])backtrace(i - 1, j - 1, path1, path2)path1.pop()path2.pop()print(dp[maxi][maxj])backtrace(maxi, maxj, [], [])return dp[maxi][maxj]PAM250 = """A C D E F G H I K L M N P Q R S T V W YA 2 -2 0 0 -3 1 -1 -1 -1 -2 -1 0 1 0 -2 1 1 0 -6 -3C -2 12 -5 -5 -4 -3 -3 -2 -5 -6 -5 -4 -3 -5 -4 0 -2 -2 -8 0D 0 -5 4 3 -6 1 1 -2 0 -4 -3 2 -1 2 -1 0 0 -2 -7 -4E 0 -5 3 4 -5 0 1 -2 0 -3 -2 1 -1 2 -1 0 0 -2 -7 -4F -3 -4 -6 -5 9 -5 -2 1 -5 2 0 -3 -5 -5 -4 -3 -3 -1 0 7G 1 -3 1 0 -5 5 -2 -3 -2 -4 -3 0 0 -1 -3 1 0 -1 -7 -5H -1 -3 1 1 -2 -2 6 -2 0 -2 -2 2 0 3 2 -1 -1 -2 -3 0I -1 -2 -2 -2 1 -3 -2 5 -2 2 2 -2 -2 -2 -2 -1 0 4 -5 -1K -1 -5 0 0 -5 -2 0 -2 5 -3 0 1 -1 1 3 0 0 -2 -3 -4L -2 -6 -4 -3 2 -4 -2 2 -3 6 4 -3 -3 -2 -3 -3 -2 2 -2 -1M -1 -5 -3 -2 0 -3 -2 2 0 4 6 -2 -2 -1 0 -2 -1 2 -4 -2N 0 -4 2 1 -3 0 2 -2 1 -3 -2 2 0 1 0 1 0 -2 -4 -2P 1 -3 -1 -1 -5 0 0 -2 -1 -3 -2 0 6 0 0 1 0 -1 -6 -5Q 0 -5 2 2 -5 -1 3 -2 1 -2 -1 1 0 4 1 -1 -1 -2 -5 -4R -2 -4 -1 -1 -4 -3 2 -2 3 -3 0 0 0 1 6 0 -1 -2 2 -4S 1 0 0 0 -3 1 -1 -1 0 -3 -2 1 1 -1 0 2 1 -1 -2 -3T 1 -2 0 0 -3 0 -1 0 0 -2 -1 0 0 -1 -1 1 3 0 -5 -3V 0 -2 -2 -2 -1 -1 -2 4 -2 2 2 -2 -1 -2 -2 -1 0 4 -6 -2W -6 -8 -7 -7 0 -7 -3 -5 -3 -2 -4 -4 -6 -5 2 -2 -5 -6 17 0Y -3 0 -4 -4 7 -5 0 -1 -4 -1 -2 -2 -5 -4 -4 -3 -3 -2 0 10"""head, *nxt = (line for line in PAM250.splitlines() if line)table = [[0] * 26 for _ in range(26)]# 处理表头head = head.split()# 处理剩余部分for row in nxt:rowBase, *nums = row.split()for colBase, num in zip(head, nums):table[chr2idx(colBase)][chr2idx(rowBase)] = int(num)# print(table)fasta = """>Rosalind_80MEANLYPRTEINSTRING>Rosalind_21PLEASANTLYEINSTEIN"""import reseqs = [seq.replace('\n', '') for seq in re.split(r'>.*', fasta) if seq.replace('\n', '')]print(seqs)Solution().localAlignment(*seqs, table)
输出结果:
23LYPRTEINSTRINLY---EINSTEIN
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | L | E | A | S | A | N | T | L | Y | E | I | N | S | T | E | I | N | |||
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 1 | M | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 0 |
| 2 | E | 0 | 0 | 0 | 8 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 4 | 0 | 3 | 0 | 0 | 4 | 0 | 3 |
| 3 | A | 0 | 1 | 0 | 3 | 10 | 5 | 2 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | 4 | 1 | 0 | 3 | 0 |
| 4 | N | 0 | 0 | 0 | 1 | 5 | 11 | 6 | 4 | 0 | 0 | 0 | 1 | 0 | 5 | 1 | 4 | 2 | 0 | 5 |
| 5 | L | 0 | 0 | 6 | 1 | 0 | 6 | 9 | 4 | 2 | 6 | 1 | 0 | 3 | 0 | 2 | 0 | 1 | 4 | 0 |
| 6 | Y | 0 | 0 | 1 | 2 | 0 | 1 | 4 | 7 | 2 | 1 | 16 | 11 | 6 | 1 | 0 | 0 | 0 | 0 | 2 |
| 7 | P | 0 | 6 | 1 | 0 | 3 | 1 | 2 | 4 | 7 | 2 | 11 | 15 | 10 | 6 | 2 | 0 | 0 | 0 | 0 |
| 8 | R | 0 | 1 | 3 | 0 | 0 | 3 | 0 | 2 | 3 | 4 | 6 | 10 | 13 | 10 | 6 | 1 | 0 | 0 | 0 |
| 9 | T | 0 | 0 | 0 | 3 | 1 | 1 | 4 | 0 | 5 | 1 | 1 | 6 | 10 | 13 | 11 | 9 | 4 | 0 | 0 |
| 10 | E | 0 | 0 | 0 | 4 | 3 | 1 | 1 | 5 | 0 | 2 | 0 | 5 | 5 | 11 | 13 | 11 | 13 | 8 | 3 |
| 11 | I | 0 | 0 | 2 | 0 | 3 | 2 | 0 | 0 | 5 | 2 | 1 | 0 | 10 | 6 | 10 | 13 | 9 | 18 | 13 |
| 12 | N | 0 | 0 | 0 | 3 | 0 | 4 | 2 | 2 | 0 | 2 | 0 | 2 | 5 | 12 | 7 | 10 | 14 | 13 | 20 |
| 13 | S | 0 | 1 | 0 | 0 | 4 | 2 | 5 | 3 | 3 | 0 | 0 | 0 | 1 | 7 | 14 | 9 | 10 | 13 | 15 |
| 14 | T | 0 | 0 | 0 | 0 | 1 | 5 | 3 | 5 | 6 | 1 | 0 | 0 | 0 | 2 | 9 | 17 | 12 | 10 | 13 |
| 15 | R | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 3 | 4 | 3 | 0 | 0 | 0 | 0 | 4 | 12 | 16 | 11 | 10 |
| 16 | I | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 3 | 6 | 2 | 0 | 5 | 0 | 0 | 7 | 11 | 21 | 16 |
| 17 | N | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 2 | 1 | 1 | 4 | 3 | 0 | 7 | 2 | 2 | 8 | 16 | 23 |
| 18 | G | 0 | 0 | 0 | 0 | 4 | 1 | 2 | 0 | 2 | 0 | 0 | 4 | 0 | 2 | 8 | 3 | 3 | 11 | 18 |
23MEANLYPRTEINSTRINLEASANTLYEINSTEIN
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | L | E | A | S | A | N | T | L | Y | E | I | N | S | T | E | I | N | |||
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 1 | M | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 0 |
| 2 | E | 0 | 0 | 0 | 8 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 4 | 0 | 3 | 0 | 0 | 4 | 0 | 3 |
| 3 | A | 0 | 1 | 0 | 3 | 10 | 5 | 2 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | 4 | 1 | 0 | 3 | 0 |
| 4 | N | 0 | 0 | 0 | 1 | 5 | 11 | 6 | 4 | 0 | 0 | 0 | 1 | 0 | 5 | 1 | 4 | 2 | 0 | 5 |
| 5 | L | 0 | 0 | 6 | 1 | 0 | 6 | 9 | 4 | 2 | 6 | 1 | 0 | 3 | 0 | 2 | 0 | 1 | 4 | 0 |
| 6 | Y | 0 | 0 | 1 | 2 | 0 | 1 | 4 | 7 | 2 | 1 | 16 | 11 | 6 | 1 | 0 | 0 | 0 | 0 | 2 |
| 7 | P | 0 | 6 | 1 | 0 | 3 | 1 | 2 | 4 | 7 | 2 | 11 | 15 | 10 | 6 | 2 | 0 | 0 | 0 | 0 |
| 8 | R | 0 | 1 | 3 | 0 | 0 | 3 | 0 | 2 | 3 | 4 | 6 | 10 | 13 | 10 | 6 | 1 | 0 | 0 | 0 |
| 9 | T | 0 | 0 | 0 | 3 | 1 | 1 | 4 | 0 | 5 | 1 | 1 | 6 | 10 | 13 | 11 | 9 | 4 | 0 | 0 |
| 10 | E | 0 | 0 | 0 | 4 | 3 | 1 | 1 | 5 | 0 | 2 | 0 | 5 | 5 | 11 | 13 | 11 | 13 | 8 | 3 |
| 11 | I | 0 | 0 | 2 | 0 | 3 | 2 | 0 | 0 | 5 | 2 | 1 | 0 | 10 | 6 | 10 | 13 | 9 | 18 | 13 |
| 12 | N | 0 | 0 | 0 | 3 | 0 | 4 | 2 | 2 | 0 | 2 | 0 | 2 | 5 | 12 | 7 | 10 | 14 | 13 | 20 |
| 13 | S | 0 | 1 | 0 | 0 | 4 | 2 | 5 | 3 | 3 | 0 | 0 | 0 | 1 | 7 | 14 | 9 | 10 | 13 | 15 |
| 14 | T | 0 | 0 | 0 | 0 | 1 | 5 | 3 | 5 | 6 | 1 | 0 | 0 | 0 | 2 | 9 | 17 | 12 | 10 | 13 |
| 15 | R | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 3 | 4 | 3 | 0 | 0 | 0 | 0 | 4 | 12 | 16 | 11 | 10 |
| 16 | I | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 3 | 6 | 2 | 0 | 5 | 0 | 0 | 7 | 11 | 21 | 16 |
| 17 | N | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 2 | 1 | 1 | 4 | 3 | 0 | 7 | 2 | 2 | 8 | 16 | 23 |
| 18 | G | 0 | 0 | 0 | 0 | 4 | 1 | 2 | 0 | 2 | 0 | 0 | 4 | 0 | 2 | 8 | 3 | 3 | 11 | 18 |

