The Secondary and Tertiary Structures of DNA
| In “Counting DNA Nucleotides”, we introduced nucleic acids, and we saw that the primary structure of a nucleic acid is determined by the ordering of its nucleobases along the sugar-phosphate backbone that constitutes the bonds of the nucleic acid polymer. Yet primary structure tells us nothing about the larger, 3-dimensional shape of the molecule, which is vital for a complete understanding of nucleic acids.
The search for a complete chemical structure of nucleic acids was central to molecular biology research in the mid-20th Century, culminating in 1953 with a publication in Nature of fewer than 800 words by James Watson and Francis Crick. Consolidating a high resolution X-ray image created by Rosalind Franklin and Raymond Gosling with a number of established chemical results, Watson and Crick proposed the following structure for DNA:
1. The DNA molecule is made up of two strands, running in opposite directions.
1. Each base bonds to a base in the opposite strand. Adenine always bonds with thymine, and cytosine always bonds with guanine; the complement of a base is the base to which it always bonds; see Figure 1.
1. The two strands are twisted together into a long spiral staircase structure called a double helix; see Figure 2.
Because they dictate how bases from different strands interact with each other, (1) and (2) above compose the secondary structure of DNA. (3) describes the 3-dimensional shape of the DNA molecule, or its tertiary structure.
In light of Watson and Crick’s model, the bonding of two complementary bases is called a base pair (bp). Therefore, the length of a DNA molecule will commonly be given in bp instead of nt. By complementarity, once we know the order of bases on one strand, we can immediately deduce the sequence of bases in the complementary strand. These bases will run in the opposite order to match the fact that the two strands of DNA run in opposite directions. | 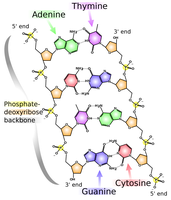
Figure 1. Base pairing across the two strands of DNA.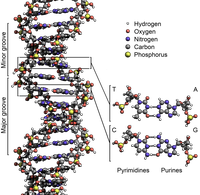
Figure 2. The double helix of DNA on the molecular scale. |
| —- | :—-: |
Problem
In DNA strings, symbols ‘A’ and ‘T’ are complements of each other, as are ‘C’ and ‘G’.
The reverse complement of a DNA string s is the string sc formed by reversing the symbols of s, then taking the complement of each symbol (e.g., the reverse complement of “GTCA” is “TGAC”).
Given: A DNA string s of length at most 1000 bp.
Return: The reverse complement sc of s.
Sample Dataset
AAAACCCGGT
Sample Output
ACCGGGTTTT
测试用例集:
TGTCTCGTAGCTGTGAGGCATGTATAAACTAACTTATCAGGCCAGGAAGAGGCCTGTAACTGTATGGCTTGATTCCCAGAAACAGGTTAGTAAATCGTAGGTACTGCGTCCTGTTACTCCGAATAACTGGAGGTTATCTGCCTTAGTAGGTAGTGCTCCTGCAGCACGCCGCGCATAGACTTAGAGTGGAGTCCTCTAGGGTGTACGTGTGATCGGGTTAGACTACGTGGTTTAGATACCCGAGGTGTAGCAAGCTTTTGGTCTAACGATCACACACCTCTTAGCACGGCATCACCACTGAGATATGACTGCGGGCAGTGCCTCAAAAATGTTACAACCAAGCGGAATGGAGTACTTCAACCCTAGTAATTGCGTCAAGTTTGGTGGAGTTTCGTTTAGCGCTACGTGTCGGGACGATGAGGTACTGTGGGAACCGTAGTTATACCGCTTGGCCGGATTTTGGGACAGTATAAAGGGAGACCCGAGGAGTCGATAACGTTAGCTATAGTTACAGCGCCGACCAGCGCATGGGGCTGCGTTTGACTACTCCCTCGGAGAAAGCAATACGTGCAGTTTTCACCAACTAAATCACCAGAATACGGAGTCGACAGATTGCGAAGGCTTGAAGTGGATCGCTGCCACGTCGGGGACGCTGTATCAATATTGGCAAGGGTGCCCTTACGATCACGTTAAATCCTACCAGAGCTGTCCGGCATAACCCTTAGTGGCTCATCGTAGATTATGCACGATGGTCCGACTTCCACCGGTGAACCGCCAGTAGAGAAAAGTGACTTAGCGGCACAGC
解题思路
本题关键是要反向互补,也就是输出要倒着来!怎么办呢?
int main() { char seqs[N]; int idx = 0; // 1.字符 IO char base = getchar(); while (base != ‘\n’) { seqs[idx++] = base; base = getchar(); } // 2.倒置输出 for (int i = idx - 1; i >= 0; i—) { switch (seqs[i]) { case ‘A’: putchar(‘T’); break; case ‘T’: putchar(‘A’); break; case ‘C’: putchar(‘G’); break; case ‘G’: putchar(‘C’); break; } } printf(“\n”); return 0; }
**编译运行**:```shell(base) b12@PC:~/ROSALIND/Complementing_a_Strand_of_DNA$ gcc -Wall ./complementDNA.c -o ./complementDNA(base) b12@PC:~/ROSALIND/Complementing_a_Strand_of_DNA$ ./complementDNATGTCTCGTAGCTGTGAGGCATGTATAAACTAACTTATCAGGCCAGGAAGAGGCCTGTAACTGTATGGCTTGATTCCCAGAAACAGGTTAGTAAATCGTAGGTACTGCGTCCTGTTACTCCGAATAACTGGAGGTTATCTGCCTTAGTAGGTAGTGCTCCTGCAGCACGCCGCGCATAGACTTAGAGTGGAGTCCTCTAGGGTGTACGTGTGATCGGGTTAGACTACGTGGTTTAGATACCCGAGGTGTAGCAAGCTTTTGGTCTAACGATCACACACCTCTTAGCACGGCATCACCACTGAGATATGACTGCGGGCAGTGCCTCAAAAATGTTACAACCAAGCGGAATGGAGTACTTCAACCCTAGTAATTGCGTCAAGTTTGGTGGAGTTTCGTTTAGCGCTACGTGTCGGGACGATGAGGTACTGTGGGAACCGTAGTTATACCGCTTGGCCGGATTTTGGGACAGTATAAAGGGAGACCCGAGGAGTCGATAACGTTAGCTATAGTTACAGCGCCGACCAGCGCATGGGGCTGCGTTTGACTACTCCCTCGGAGAAAGCAATACGTGCAGTTTTCACCAACTAAATCACCAGAATACGGAGTCGACAGATTGCGAAGGCTTGAAGTGGATCGCTGCCACGTCGGGGACGCTGTATCAATATTGGCAAGGGTGCCCTTACGATCACGTTAAATCCTACCAGAGCTGTCCGGCATAACCCTTAGTGGCTCATCGTAGATTATGCACGATGGTCCGACTTCCACCGGTGAACCGCCAGTAGAGAAAAGTGACTTAGCGGCACAGCGCTGTGCCGCTAAGTCACTTTTCTCTACTGGCGGTTCACCGGTGGAAGTCGGACCATCGTGCATAATCTACGATGAGCCACTAAGGGTTATGCCGGACAGCTCTGGTAGGATTTAACGTGATCGTAAGGGCACCCTTGCCAATATTGATACAGCGTCCCCGACGTGGCAGCGATCCACTTCAAGCCTTCGCAATCTGTCGACTCCGTATTCTGGTGATTTAGTTGGTGAAAACTGCACGTATTGCTTTCTCCGAGGGAGTAGTCAAACGCAGCCCCATGCGCTGGTCGGCGCTGTAACTATAGCTAACGTTATCGACTCCTCGGGTCTCCCTTTATACTGTCCCAAAATCCGGCCAAGCGGTATAACTACGGTTCCCACAGTACCTCATCGTCCCGACACGTAGCGCTAAACGAAACTCCACCAAACTTGACGCAATTACTAGGGTTGAAGTACTCCATTCCGCTTGGTTGTAACATTTTTGAGGCACTGCCCGCAGTCATATCTCAGTGGTGATGCCGTGCTAAGAGGTGTGTGATCGTTAGACCAAAAGCTTGCTACACCTCGGGTATCTAAACCACGTAGTCTAACCCGATCACACGTACACCCTAGAGGACTCCACTCTAAGTCTATGCGCGGCGTGCTGCAGGAGCACTACCTACTAAGGCAGATAACCTCCAGTTATTCGGAGTAACAGGACGCAGTACCTACGATTTACTAACCTGTTTCTGGGAATCAAGCCATACAGTTACAGGCCTCTTCCTGGCCTGATAAGTTAGTTTATACATGCCTCACAGCTACGAGACA
void solve() { char base = getchar(); if (‘\n’ == base) return; // 2.递归出口 solve(); // 1.先递归到最深 // 3. 回溯倒置打印 if (‘A’ == base) { putchar(‘T’); } else if (‘T’ == base) { putchar(‘A’); } else if (‘C’ == base) { putchar(‘G’); } else if (‘G’ == base) { putchar(‘C’); } }
int main() { solve(); printf(“\n”); return 0; }
编译运行:
(base) b12@PC:~/ROSALIND/Complementing_a_Strand_of_DNA$ gcc -Wall ./complementDNA.c -o ./complementDNA (base) b12@PC:~/ROSALIND/Complementing_a_Strand_of_DNA$ ./complementDNA TTCCGACAGTGAAGCCGACGGTCCTGCGTATGAATTACCAGTACGTTGTCCCCATCCCGGCTTATCTCGTCTACACTAGATTTCTAGTGTGAGCCTTCACAGCTCCTCTACTTAGTGAATGGTTAATTGGCCCTCAACGAGGAAGCTACAGCAATAACAAGTAATCTGTAGTGGGACGGTCCCGAACAGGTAGACGATGTTGAAGCATTAAACCACTATCTGAGGAGCACCTAAGCCTTATGCCCACCACGATACAGCAACGTGCCAGAACTGATGGACTAGGCCGGCGAGGACACGTGACATGACGTCCTATCAACACTCGGATCTGTCGTTAATCGAAGGTGAAAGAATCGCCCGAGGGAGCACTTGCCTTCAAGACGAATTTGATTCAGGTACTTACAGACATACACTCGAATGGGAAGGTGCCTAGGTTCGTTGGACCGACGTTTCCGAATTACTTTGAACCCACCCGGTCGAGATAGGCCATTATCTGACGGGGTTCGTCTAGCAAGGGAAATCCTAGGGCCAGCTCGTGAGTAAGTTTATTCAAGCAACAGACGGGATCAAGGAATCCTCAATCACAGCTGCTACTCTGTGATTTTTCGTTAACCAGTCTTTAGGCTACTATTGCGCGCGCGGTACATGTCCGTGCGCTAGTTAGAGACAGTCCAGGGCGCGTCGGGTCATGTCCCGACCATGACAAAATGACTTACTTTAGTGGAACAGTCATTTTACTATCGTGGCCTGATAAGTCAGGGAGAAGGCGCAAGACTTTGCTATTTGGTACTCCATGTTGGTGACTCATGTCTGATGTGCCGGACCTATAGTGCAGACTGATCCATTGTATTGGGTGGTAGTCGCTGCGT ACGCAGCGACTACCACCCAATACAATGGATCAGTCTGCACTATAGGTCCGGCACATCAGACATGAGTCACCAACATGGAGTACCAAATAGCAAAGTCTTGCGCCTTCTCCCTGACTTATCAGGCCACGATAGTAAAATGACTGTTCCACTAAAGTAAGTCATTTTGTCATGGTCGGGACATGACCCGACGCGCCCTGGACTGTCTCTAACTAGCGCACGGACATGTACCGCGCGCGCAATAGTAGCCTAAAGACTGGTTAACGAAAAATCACAGAGTAGCAGCTGTGATTGAGGATTCCTTGATCCCGTCTGTTGCTTGAATAAACTTACTCACGAGCTGGCCCTAGGATTTCCCTTGCTAGACGAACCCCGTCAGATAATGGCCTATCTCGACCGGGTGGGTTCAAAGTAATTCGGAAACGTCGGTCCAACGAACCTAGGCACCTTCCCATTCGAGTGTATGTCTGTAAGTACCTGAATCAAATTCGTCTTGAAGGCAAGTGCTCCCTCGGGCGATTCTTTCACCTTCGATTAACGACAGATCCGAGTGTTGATAGGACGTCATGTCACGTGTCCTCGCCGGCCTAGTCCATCAGTTCTGGCACGTTGCTGTATCGTGGTGGGCATAAGGCTTAGGTGCTCCTCAGATAGTGGTTTAATGCTTCAACATCGTCTACCTGTTCGGGACCGTCCCACTACAGATTACTTGTTATTGCTGTAGCTTCCTCGTTGAGGGCCAATTAACCATTCACTAAGTAGAGGAGCTGTGAAGGCTCACACTAGAAATCTAGTGTAGACGAGATAAGCCGGGATGGGGACAACGTACTGGTAATTCATACGCAGGACCGTCGGCTTCACTGTCGGAA ```

