如何筛选多因素cox组合?
https://blog.csdn.net/DJXtxdy/article/details/119011074
An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics
:::info
Cell (IF: 41.58; Q1). 2018 Apr 5;173(2):400-416.e11. doi: 10.1016/j.cell.2018.02.052.
Cited by: 1034
Publish Year: 2018
:::
SUMMARY
For a decade, The Cancer Genome Atlas (TCGA) program collected clinicopathologic annotation data along with multi-platform molecular profiles of more than 11,000 human tumors across 33 different cancer types. TCGA clinical data contain key features representing the democratized nature of the data collection process. To ensure proper use of this large clinical dataset associated with genomic features, we developed a standardized dataset named the TCGA Pan-Cancer Clinical Data Resource (TCGA-CDR), which includes four major clinical outcome endpoints. In addition to detailing major challenges and statistical limitations encountered during the effort of integrating the acquired clinical data, we present a summary that includes endpoint usage recommendations for each cancer type. These TCGA-CDR findings appear to be consistent with cancer genomics studies independent of the TCGA effort and provide opportunities for investigating cancer biology using clinical correlates at an unprecedented scale.
Graphical abstract
In Brief Analysis of clinicopathologic annotations for over 11,000 cancer patients in the TCGA program leads to the generation of TCGA Clinical Data Resource, which provides recommendations of clinical outcome endpoint usage for 33 cancer types.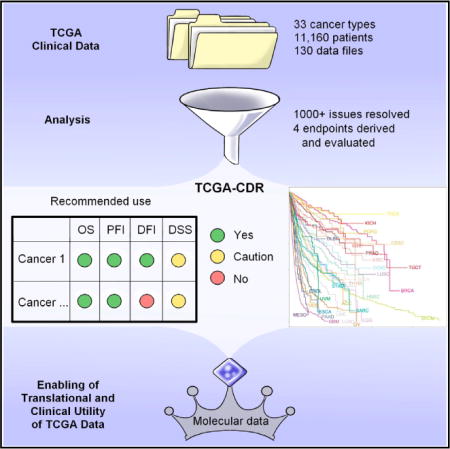
INTRODUCTION
The purpose of The Cancer Genome Atlas (TCGA) project was to establish a coordinated(协调一致的) team science effort to comprehensively characterize the molecular events in primary cancers and to provide these data to the public for use by researchers around the world. TCGA started in 2006 with a 3-year pilot project(试点项目) focusing on glioblastoma multiforme (GBM,多形性成胶质细胞瘤), lung squamous cell carcinoma (LUSC,肺鳞癌), and ovarian serious cystadenocarcinoma (OV,卵巢严重膀胱瘤癌), followed by the execution of the full project from 2009 to 2015. By the end of this 10-year project, TCGA network investigators had characterized the molecular landscape of tumors from 11,160 patients across 33 cancer types and defined their many molecular subtypes. The quantity and quality of TCGA molecular data have been lauded(laud美: [lɔd] 英: [lɔːd] n.赞美;赞美歌;松歌;v.称赞) by a large number of scientists, and these data have resulted in studies that have significantly advanced our understanding of cancer biology, as documented in dozens of highly cited published TCGA marker and companion papers, including those for GBM, OV, and breast, lung, prostate(prostate美: [ˈprɑˌsteɪt] 英: [ˈprɒˌsteɪt] n.前列腺), bladder(bladder美: [ˈblædər] 英: [ˈblædə(r)] n.膀胱;皮囊), and other individual cancers (Cancer Genome Atlas Network, 2012, 2015; The Cancer Genome Atlas Research Network, 2008, 2011, 2012, 2014, 2015; Cancer Genome Atlas Research Network et al., 2017). TCGA data also make possible studies that compare and contrast multiple cancer types with the goal of identifying common themes that transcend the tissue of origin and may inform precision oncology (Hoadley et al., 2014). In addition, numerous independent investigators have used TCGA as a resource to support their own studies and to help interpret molecular testing of individual patients in a clinical setting (Huo et al., 2017; Verhaak et al., 2010). However, obtaining comprehensive clinical annotation was neither a primary program objective nor a practical possibility, given the worldwide scope and severe time constraints for sample accrual goals determined at the time of TCGA program initiation and funding. The incomplete annotation of patient outcome and treatment data associated with each TCGA-acquired sample, with its relatively short-term clinical follow-up interval, has been noted by the research community (Hoadley et al., 2014; Huo et al., 2017). The limitations of the existing clinical dataset, associated with an otherwise rich body of genomic and molecular analyses available across all TCGA tumor types, compels(compel美: [kəmˈpel] 英: [kəm’pel] v.强迫;协迫;使不得不;迫使(服从,沉默等)) thorough and systematic curation and evaluation of those clinical endpoints and other clinical features associated with each TCGA tumor so that the scientific community can optimize the translational relevance of the tumor-specific genomic and pathway conclusions drawn from the TCGA program and its pan-cancer analyses. It is also important to demonstrate that the conclusions drawn from this newly curated TCGA pan-cancer clinical data resource have translational validity with respect to both patient prognosis and outcome parameters.
In clinical studies, 5-year or 10-year benchmark survival rates are often calculated to convey prognostic information or to compare treatment effects. These survival rates may be based on progression or mortality events with or without disease specificity. For each endpoint, it is very important to have a sufficiently long follow-up time to capture the events of interest, and the minimum follow-up time needed depends on both the aggressiveness of the disease and the type of endpoint (Tai et al., 2005).
Overall survival (OS) is an important endpoint, with the advantage that there is minimal ambiguity in defining an OS event (Hudis et al., 2007; Punt et al., 2007); the patient is either alive or dead. However, using OS as an endpoint may weaken a clinical study as deaths because of non-cancer causes do not necessarily reflect tumor biology, aggressiveness, or responsiveness to therapy. Using OS or disease-specific survival (DSS,疾病相关存活率) demands longer follow-up times; thus, in many clinical trials, disease-free interval (DFI,无病生存期的长短;无病间歇期;无病间隔期间) or progression-free interval (PFI,无瘤间期;缓解期) are used (Hudis et al., 2007; Punt et al., 2007; https://wiki.nci.nih.gov/plugins/servlet/mobile#content/view/24279961). The minimum follow-up time for these endpoints is shorter because patients generally develop disease recurrence or progression before dying of their disease. Selection of a specific survival endpoint also depends on the study goal. For example, a clinical trial testing the effect of a drug’s ability to delay or prevent cancer progression would use PFI as the most appropriate endpoint.(短期疗效) With specific regard to the analysis of available TCGA clinical data, it is important to realize that short-term clinical follow-up intervals favor outcome analyses in more aggressive cancer types, which are likely to observe events within a couple of years. Studies with less aggressive cancer types, in which patients relapse only after many years or even decades, may not observe enough events during their follow-up intervals to support reliable outcome determinations. The intent of this analysis is to examine the relative strengths and weaknesses of the TCGA pan-cancer clinical outcome measures to guide future analyses and avoid pitfalls(pitfall美: [ˈpɪtˌfɔl] 英: [ˈpɪtˌfɔːl] n.陷阱;诱惑;圈套;隐藏的危险【网络】缺陷;易犯的错误;这方式有陷阱) such as insufficient follow-up intervals.
To our knowledge, there has been no systematic attempt to analyze the TCGA clinical data and derive acceptable outcome endpoints across all 33 TCGA cancer types involving 11,160 patients or to assess the adequacy of the clinical follow-up interval for each survival endpoint test. Here we present curated and filtered clinical and survival outcome data as a newly integrated resource for the entire scientific community, describe how problems encountered while analyzing these data were resolved, and what pitfalls researchers should be aware of when using these data for future correlative and survival studies. Based on our comprehensive clinical review, we also provide scoring recommendations for appropriate future use and tumor-specific endpoint selection. The resulting compendium(compendium美: [kəmˈpendiəm] 英: [kəm’pendiəm] n.(尤指书中某题材事实、图画及照片的)汇编【网络】概略;纲要;概要) of curated data is now presented as the TCGA Pan-Cancer Clinical Data Resource (TCGA-CDR) for public access and future translational cancer research.
RESULTS
The TCGA clinical data were downloaded from the data portal of the Genomic Data Commons (GDC), where all TCGA molecular data are also available (https://gdc-portal.nci.nih.gov/legacy-archive/). The same TCGA barcode structure is used for both clinical data and molecular data, enabling integrated analysis of patient-based clinical data and sample-based molecular data.
Cohort Characteristics
Figure 1A shows a flowchart of the methods for clinical data integration and analysis as well as derivation and evaluation of 4 major clinical outcome endpoints. We processed 33 initial enrollment data files and 97 follow-up data files for 11,160 patients across 33 cancer types. Table 1 shows the basic characteristics of each TCGA cohort. Primary tumor samples, not metastatic, were typically selected in each cohort for molecular characterization, with the exception of the skin cutaneous melanoma (SKCM,皮肤皮肤黑色素瘤) study, which allowed both. A very limited number of metastatic tumors with matching primary tumors was also studied for other cancer types. Individual patients’ detailed data are provided in Table S1, tab TCGA-CDR, and problems we identified when processing this dataset and the solutions we developed are described in the STAR Methods.
Table 3、Assessment and Recommended Use of the Endpoints of OS, PFI, DFI, and DSS
| Type | N | OS (Accurately Defined) | PFI (Accurately Defined) | DFI (Accurately Defined) | DSS (Approximately Defined) | Explanation/Caution | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Use | Event | Censored | Use | Event | Censored | Use | Event | Censored | Use | Event | Censored | |||
| BRCA | 1097 | ✓* | 151 | 946 | ✓ | 145 | 952 | ✓ | 84 | 869 | ✓ App.* | 83 | 995 | need a longer follow-up for OS and DSS |
✓recommended for use (passed at least passed one of the 3 tests in step 1 and the supplemental checks in step 2 as described in the STAR Methods);
×not recommended for use;
*caution, see the explanation/caution column; app., approximate; acc., accurate.
Validation and Application Examples
In breast cancer studies, patients with estrogen receptor-negative (ER−) tumors have worse clinical survival outcomes compared with those with ER-positive (ER+) tumors. To evaluate the derived clinical endpoints, we compared the survival of patients with these two types of tumors using OS, PFI, DFI, and DSS, respectively (Figures 2A–2D; plots truncated at 10-year follow-up time, but analyses were conducted using the whole dataset following Huo et al., 2017). Univariate analyses showed that TCGA breast cancer patients with ER+ tumors had better survival than patients with ER− tumors when using PFI (p = 0.005) and DFI (p = 0.001) as clinical endpoints, but there was no sufficient evidence of a difference when using OS as the endpoint (p = 0.097). We also noticed that there was a significant difference in (approximated) DSS (p = 0.009), demonstrating the potential value of this estimated endpoint. As noted in Table 3, although we caution against using breast invasive carcinoma (BRCA) data to determine OS and DSS, the above findings validate our recommended use of PFI and DFI as suitable endpoints for specific types of breast cancer molecular studies.

