Motif Implies Function**
As mentioned in “Translating RNA into Protein”, proteins perform every practical function in the cell. A structural and functional unit of the protein is a protein domain: in terms of the protein’s primary structure, the domain is an interval of amino acids that can evolve and function independently.
Each domain usually corresponds to a single function of the protein (e.g., binding the protein to DNA, creating or breaking specific chemical bonds, etc.). Some proteins, such as myoglobin and the Cytochrome complex, have only one domain, but many proteins are multifunctional and therefore possess several domains. It is even possible to artificially fuse different domains into a protein molecule with definite properties, creating a chimeric protein.
| Just like species, proteins can evolve, forming homologous groups called protein families. Proteins from one family usually have the same set of domains, performing similar functions; see Figure 1. A component of a domain essential for its function is called a motif, a term that in general has the same meaning as it does in nucleic acids, although many other terms are also used (blocks, signatures, fingerprints, etc.) Usually protein motifs are evolutionarily conservative, meaning that they appear without much change in different species. Proteins are identified in different labs around the world and gathered into freely accessible databases. A central repository for protein data is UniProt, which provides detailed protein annotation, including function description, domain structure, and post-translational modifications. UniProt also supports protein similarity search, taxonomy analysis, and literature citations. |
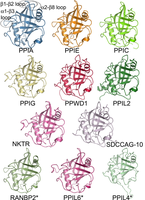 Figure 1. The human cyclophilin family, as represented by the structures of the isomerase domains of some of its members. |
|---|---|
Problem
To allow for the presence of its varying forms, a protein motif is represented by a shorthand as follows: [XY] means “either X or Y” and {X} means “any amino acid except X.” For example, the N-glycosylation motif is written as N{P}[ST]{P}.
You can see the complete description and features of a particular protein by its access ID “uniprot_id” in the UniProt database, by inserting the ID number into
http://www.uniprot.org/uniprot/uniprot_id
Alternatively, you can obtain a protein sequence in FASTA format by following
http://www.uniprot.org/uniprot/uniprot_id.fasta
For example, the data for protein B5ZC00 can be found at http://www.uniprot.org/uniprot/B5ZC00.
Given: At most 15 UniProt Protein Database access IDs.
Return: For each protein possessing the N-glycosylation motif, output its given access ID followed by a list of locations in the protein string where the motif can be found.
Sample Dataset
A2Z669B5ZC00P07204_TRBM_HUMANP20840_SAG1_YEAST
Sample Output
B5ZC0085 118 142 306 395P07204_TRBM_HUMAN47 115 116 382 409P20840_SAG1_YEAST79 109 135 248 306 348 364 402 485 501 614
Solution
本例涉及到爬虫的入门知识和文本处理,但是真的很简单。爬虫返回的文档是
'>sp|A2Z669|CSPLT_ORYSI CASP-like protein 5A2 OS=Oryza sativa subsp. indica OX=39946 GN=OsI_33147 PE=3 SV=1\nMRASRPVVHPVEAPPPAALAVAAAAVAVEAGVGAGGGAAAHGGENAQPRGVRMKDPPGAP\nGTPGGLGLRLVQAFFAAAALAVMASTDDFPSVSAFCYLVAAAILQCLWSLSLAVVDIYAL\nLVKRSLRNPQAVCIFTIGDGITGTLTLGAACASAGITVLIGNDLNICANNHCASFETATA\nMAFISWFALAPSCVLNFWSMASR\n'
即标准的 fasta 格式,第一行就是序列 ID ,这行可以不用管他,然后只需要后面几行的序列组成一个字符串,可见只需要把 \n 给删除掉就行了。
对于本题只需要正则表达式实在简单,直接使用 N[^P][ST][^P],根本不用考虑 overlapping 匹配。因为要获得位置,所以使用 re.finditer 函数获得 re.Match object 然后调用其 start() 获得匹配索引起始位置(记住答案是要 +1)
import requests, reproteins = ['A2Z669', 'B5ZC00', 'P07204_TRBM_HUMAN', 'P20840_SAG1_YEAST']pat = re.compile(r'N[^P][ST][^P]')for protein in proteins:url = f'http://www.uniprot.org/uniprot/{protein}.fasta'seq = ''.join(requests.get(url).text.split('\n')[1:])position = [i.start() + 1 for i in re.finditer(pat, seq)]if position:print(protein)print(' '.join(map(str, position)))
拓展延申:正则表达式的前向断言!overlapping 实现(?=...)
Matches if ... matches next, but doesn’t consume any of the string. This is called a lookahead assertion. For example, Isaac (?=Asimov) will match 'Isaac ' only if it’s followed by 'Asimov'.
Positive lookahead assertion. This succeeds if the contained regular expression, represented here by ..., successfully matches at the current location, and fails otherwise. But, once the contained expression has been tried, the matching engine doesn’t advance at all; the rest of the pattern is tried right where the assertion started.
怎么说呢?就是它匹配后不会跳过它字符串!到下个位置开始,具体见下面示例
s = 'ABABA'import reprint(re.search(r'ABA', s))print(re.findall(r'ABA', s))print(list(re.finditer(r'(ABA)', s)))print(re.findall(r'(ABA)', s))# 非捕获组:overlappingprint(re.findall(r'(?=ABA)', s))print(list(re.finditer(r'(?=ABA)', s)))# 非捕获组:获取捕获内容?print(re.findall(r'(?=(ABA))', s))print(list(map(lambda x: x.group(1), re.finditer(r'(?=(ABA))', s))))
输出结果:
<re.Match object; span=(0, 3), match='ABA'>['ABA'][<re.Match object; span=(0, 3), match='ABA'>]['ABA']['', ''][<re.Match object; span=(0, 0), match=''>, <re.Match object; span=(2, 2), match=''>]['ABA', 'ABA']['ABA', 'ABA']
即 (?=...) 是一个匹配一个 () 内的表达式后,原来的匹配位置不会跑到这个串的结尾,也就是不动,还可以继续往后匹配(overlapping 原理)!但是这样可以做到覆盖匹配,却无法取出这个匹配的内容,该怎么办?那就在里面再套上一个捕获组,这样根据组的顺序命名 (?=(ABA)) ,最里面的 ABA 就是第一个组,而第二个组才是 (?=) 非捕获组,因此大功告成。
:::tips
本例最内层的捕获组虽然你知道是 ABA ,也可根据最终匹配结果进行推导,但是你万一不确定这个捕获组的内容呢?因此 () 就是一个高级的东西,里面还可以嵌套表达式啊,这样非常方便。
:::

