获取更多R语言知识,请关注公众号:医学和生信笔记
医学和生信笔记 公众号主要分享:1.医学小知识、肛肠科小知识;2.R语言和Python相关的数据分析、可视化、机器学习等;3.生物信息学学习资料和自己的学习笔记!
多时间点ROC
加载R包和数据
rm(list = ls())library(timeROC)library(survival)load(file = "../000files/timeROC.RData")
多个时间点ROC
首先看一下数据结构,对于多个时间点的ROC,需要3列数据:time, event, marker(比如你计算得到的risk score)
# 看一下画图所需的数据长什么样子,event这一列,0代表living,1代表dead,futime这一列单位是年,也可以改成# 其他的knitr::kable(df[1:10,])
| event | riskScore | futime |
|---|---|---|
| 0 | -0.2493257 | 3.0275000 |
| 0 | -0.5111057 | 1.1558333 |
| 1 | -0.2113056 | 1.8191667 |
| 0 | -0.4270568 | 1.5166667 |
| 0 | 0.2785857 | 1.3441667 |
| 1 | -0.0067608 | 0.0500000 |
| 0 | -0.3104566 | 2.3108333 |
| 0 | -0.3660733 | 1.0225000 |
| 0 | -0.2568007 | 7.5083333 |
| 0 | -0.0232976 | 0.3308333 |
str(df)## 'data.frame': 297 obs. of 3 variables:## $ event : num 0 0 1 0 0 1 0 0 0 0 ...## $ riskScore: num -0.249 -0.511 -0.211 -0.427 0.279 ...## $ futime : num 3.03 1.16 1.82 1.52 1.34 ...
# 下面是画图代码ROC <- timeROC(T=df$futime,delta=df$event,marker=df$riskScore,cause=1, #阳性结局指标数值weighting="marginal", #计算方法,默认为marginaltimes=c(1, 2, 3), #时间点,选取1年,3年和5年的生存率iid=TRUE)ROC #查看模型变量信息## Time-dependent-Roc curve estimated using IPCW (n=297, without competing risks).## Cases Survivors Censored AUC (%) se## t=1 57 203 37 71.02 3.68## t=2 66 106 125 69.23 3.94## t=3 68 74 155 65.53 4.85#### Method used for estimating IPCW:marginal#### Total computation time : 0.19 secs.# tiff("figures/time_roc.tiff",width = 12,height = 12,units = "cm",compression = "lzw", res = 500)plot(ROC,time=1, col="red", lwd=2, title = "") #time是时间点,col是线条颜色plot(ROC,time=2, col="blue", add=TRUE, lwd=2) #add指是否添加在上一张图中plot(ROC,time=3, col="orange", add=TRUE, lwd=2)#添加标签信息legend("bottomright",c(paste0("AUC at 1 year: ",round(ROC[["AUC"]][1],2)),paste0("AUC at 2 year: ",round(ROC[["AUC"]][2],2)),paste0("AUC at 3 year: ",round(ROC[["AUC"]][3],2))),col=c("red", "blue", "orange"),lty=1, lwd=2,bty = "n")
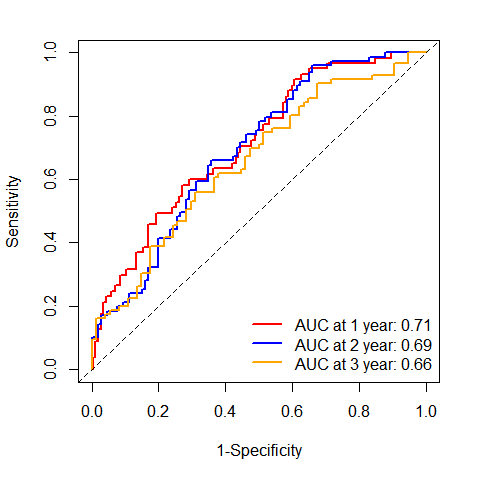
# dev.off()
多指标ROC
首先也是看一下所需要的数据结构,其中futime和event是必须的,另外的几列是你想要用来画ROC曲线图的指标,可以自己添加,在这里我使用了riskScore, gender, TNM分期。 在gender这一列,1是female,2是male,t,n,m这3列,数字代表不同的分期
knitr::kable(df2[1:10,])
| event | age | riskScore | futime | gender | t | n | m |
|---|---|---|---|---|---|---|---|
| 0 | 59 | -0.2493257 | 3.0275000 | 2 | 4 | 1 | 1 |
| 0 | 63 | -0.5111057 | 1.1558333 | 2 | 4 | 5 | 1 |
| 1 | 65 | -0.2113056 | 1.8191667 | 2 | 4 | 1 | 1 |
| 0 | 73 | -0.4270568 | 1.5166667 | 1 | 3 | 1 | 1 |
| 0 | 59 | 0.2785857 | 1.3441667 | 2 | 3 | 1 | 1 |
| 1 | 66 | -0.0067608 | 0.0500000 | 2 | 3 | 1 | 3 |
| 0 | 56 | -0.3104566 | 2.3108333 | 1 | 5 | 3 | 1 |
| 0 | 42 | -0.3660733 | 1.0225000 | 2 | 3 | 1 | 1 |
| 0 | 61 | -0.2568007 | 7.5083333 | 2 | NA | NA | 3 |
| 0 | 48 | -0.0232976 | 0.3308333 | 2 | 4 | 1 | 3 |
str(df2)## 'data.frame': 297 obs. of 8 variables:## $ event : num 0 0 1 0 0 1 0 0 0 0 ...## $ age : int 59 63 65 73 59 66 56 42 61 48 ...## $ riskScore: num -0.249 -0.511 -0.211 -0.427 0.279 ...## $ futime : num 3.03 1.16 1.82 1.52 1.34 ...## $ gender : num 2 2 2 1 2 2 1 2 2 2 ...## $ t : num 4 4 4 3 3 3 5 3 NA 4 ...## $ n : num 1 5 1 1 1 1 3 1 NA 1 ...## $ m : num 1 1 1 1 1 3 1 1 3 3 ...
# riskScore的ROC曲线ROC.risk <- timeROC(T=df2$futime,delta=df2$event,marker=df2$riskScore,cause=1,weighting="marginal",times=3,iid=TRUE)# gender的ROC曲线ROC.gender <- timeROC(T=df2$futime,delta=df2$event,marker=df2$gender,cause=1,weighting="marginal",times=3,iid=TRUE)# age的ROC曲线ROC.age <- timeROC(T=df2$futime,delta=df2$event,marker=df2$age,cause=1,weighting="marginal",times=3,iid=TRUE)# T分期的ROC曲线ROC.T <- timeROC(T=df2$futime,delta=df2$event,marker=df2$t,cause=1,weighting="marginal",times=3,iid=TRUE)# N分期的ROC曲线ROC.N <- timeROC(T=df2$futime,delta=df2$event,marker=df2$n,cause=1,weighting="marginal",times=3,iid=TRUE)# M分期的ROC曲线ROC.M <- timeROC(T=df2$futime,delta=df2$event,marker=df2$m,cause=1,weighting="marginal",times=3,iid=TRUE)# tiff("figures/多指标ROC.tiff",width = 12,height = 12,units = "cm",compression = "lzw", res = 500)plot(ROC.risk, time = 3, col="#E41A1C", lwd=2, title = "")plot(ROC.gender, time = 3, col="#A65628", lwd=2, add = T)plot(ROC.age, time = 3, col="#4DAF4A", lwd=2, add = T)plot(ROC.T, time = 3, col="#377EB8", lwd=2, add = T)plot(ROC.N, time = 3, col="#984EA3", lwd=2, add = T)plot(ROC.M, time = 3, col="#FFFF33", lwd=2, add = T)legend("bottomright",c(paste0("Risk score: ",round(ROC.risk[["AUC"]][2],2)),paste0("gender: ",round(ROC.gender[["AUC"]][2],2)),paste0("age: ",round(ROC.age[["AUC"]][2],2)),paste0("T: ",round(ROC.T[["AUC"]][2],2)),paste0("N: ",round(ROC.N[["AUC"]][2],2)),paste0("M: ",round(ROC.M[["AUC"]][2],2))),col=c("#E41A1C", "#A65628", "#4DAF4A","#377EB8","#984EA3","#FFFF33"),lty=1, lwd=2,bty = "n")
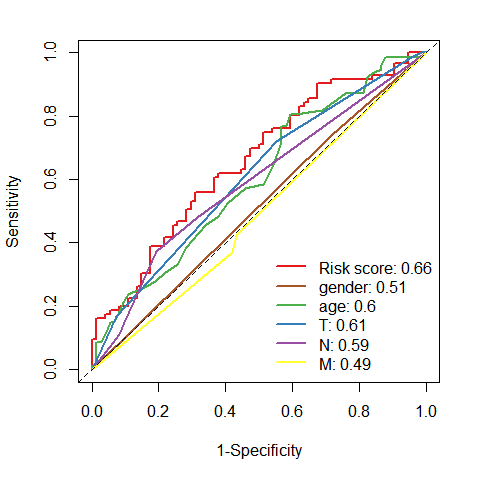
# dev.off()# "#E41A1C" "#377EB8" "#4DAF4A" "#984EA3" "#FF7F00" "#FFFF33" "#A65628" "#F781BF"
获取更多R语言知识,请关注公众号:医学和生信笔记
医学和生信笔记 公众号主要分享:1.医学小知识、肛肠科小知识;2.R语言和Python相关的数据分析、可视化、机器学习等;3.生物信息学学习资料和自己的学习笔记!

