- Chapter 22. Composable Data at Cerner
- Saving Lives with Software
- Chapter 24. Cascading
- Appendix A. Installing Apache Hadoop
- Appendix B. Cloudera’s Distribution Including Apache Hadoop
- Appendix C. Preparing the NCDC Weather Data
- Appendix D. The Old and New Java MapReduce APIs
Chapter 22. Composable Data at Cerner
Ryan Brush Micah Whitacre
Healthcare information technology is often a story of automating existing processes. This is changing. Demands to improve care quality and control its costs are growing, creating a need for better systems to support those goals. Here we look at how Cerner is using the Hadoop ecosystem to make sense of healthcare and — building on that knowledge — to help solve such problems.
From CPUs to Semantic Integration
Cerner has long been focused on applying technology to healthcare, with much of our history emphasizing electronic medical records. However, new problems required a broader approach, which led us to look into Hadoop.
In 2009, we needed to create better search indexes of medical records. This led to processing needs not easily solved with other architectures. The search indexes required expensive processing of clinical documentation: extracting terms from the documentation and resolving their relationships with other terms. For instance, if a user typed “heart disease,” we wanted documents discussing a myocardial infarction to be returned. This processing was quite expensive — it can take several seconds of CPU time for larger documents — and we wanted to apply it to many millions of documents. In short, we needed to throw a lot of CPUs at the problem, and be cost effective in the process.
Among other options, we considered a staged event-driven architecture (SEDA) approach to ingest documents at scale. But Hadoop stood out for one important need: we wanted to reprocess the many millions of documents frequently, in a small number of hours or faster. The logic for knowledge extraction from the clinical documents was rapidly improving, and we needed to roll improvements out to the world quickly. In Hadoop, this simply meant running a new version of a MapReduce job over data already in place. The process documents were then loaded into a cluster of Apache Solr servers to support application queries.
These early successes set the stage for more involved projects. This type of system and its data can be used as an empirical basis to help control costs and improve care across entire populations. And since healthcare data is often fragmented across systems and institutions, we needed to first bring in all of that data and make sense of it.
With dozens of data sources and formats, and even standardized data models subject to interpretation, we were facing an enormous semantic integration problem. Our biggest challenge was not the size of the data — we knew Hadoop could scale to our needs — but the sheer complexity of cleaning, managing, and transforming it for our needs. We needed higher-level tools to manage that complexity.
Enter Apache Crunch
Bringing together and analyzing such disparate datasets creates a lot of demands, but a few stood out: We needed to split many processing steps into modules that could easily be assembled into a sophisticated pipeline.
We needed to split many processing steps into modules that could easily be assembled into a sophisticated pipeline.
We needed to offer a higher-level programming model than raw MapReduce.
We needed to work with the complex structure of medical records, which have several hundred unique fields and several levels of nested substructures.
We explored a variety of options in this case, including Pig, Hive, and Cascading. Each of these worked well, and we continue to use Hive for ad hoc analysis, but they were unwieldy when applying arbitrary logic to our complex data structures. Then we heard of Crunch (see Chapter 18), a project led by Josh Wills that is similar to the FlumeJava system from Google. Crunch offers a simple Java-based programming model and static type checking of records — a perfect fit for our community of Java developers and the type of data we were working with.
Building a Complete Picture
Understanding and managing healthcare at scale requires significant amounts of clean, normalized, and relatable data. Unfortunately, such data is typically spread across a number of sources, making it difficult and error prone to consolidate. Hospitals, doctors’ offices, clinics, and pharmacies each hold portions of a person’s records in industrystandard formats such as CCDs (Continuity of Care Documents), HL7 (Health Level 7, a healthcare data interchange format), CSV files, or proprietary formats.
Our challenge is to take this data; transform it into a clean, integrated representation; and use it to create registries that help patients manage specific conditions, measure operational aspects of healthcare, and support a variety of analytics, as shown in Figure 22-1.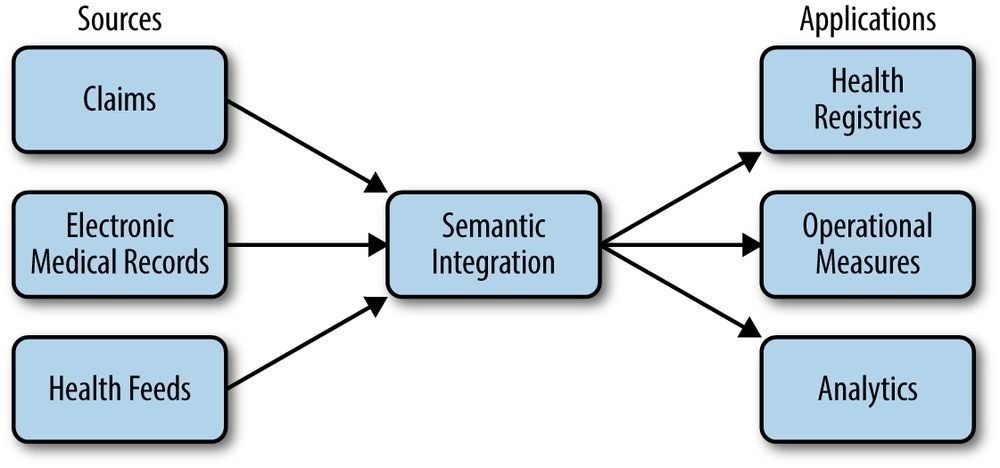
Figure 22-1. Operational data flow
An essential step is to create a clean, semantically integrated basis we can build on, which is the focus of this case study. We start by normalizing data to a common structure. Earlier versions of this system used different models, but have since migrated to Avro for storing and sharing data between processing steps. Example 22-1 shows a simplified Avro IDL to illustrate how our common structures look.
Example 22-1. Avro IDL for common data types
@namespace(“com.cerner.example”) protocol PersonProtocol {
record Demographics { string firstName; string lastName; string dob;
… }
record LabResult { string personId; string labDate; int labId; int labTypeId; int value; }
record Medication { string personId; string medicationId; string dose; string doseUnits; string frequency;
… }
record Diagnosis { string personId; string diagnosisId; string date;
… }
record Allergy { string personId; int allergyId; int substanceId;
… }
/*
Represents a person’s record from a single source.
*/
record PersonRecord { string personId;
Demographics demographics; array
. . .
} }
Note that a variety of data types are all nested in a common person record rather than in separate datasets. This supports the most common usage pattern for this data — looking at a complete record — without requiring downstream operations to do a number of expensive joins between datasets.
A series of Crunch pipelines are used to manipulate the data into a
PCollection
Table 22-1. Aggregated data
Source Person ID Person demographics Data
| Doctor’s office 12345 | Abraham Lincoln … | Diabetes diagnosis, lab results |
|---|---|---|
| Hospital 98765 | Abe Lincoln … | Flu diagnosis |
| Pharmacy 98765 | Abe Lincoln … | Allergies, medications |
| Clinic 76543 | A. Lincoln … | Lab results |
Consumers wishing to retrieve data from a collection of authorized sources call a “retriever” API that simply produces a Crunch PCollection of requested data:
Set
PCollection
RecordRetriever.getData(pipeline, sources);
This retriever pattern allows consumers to load datasets while being insulated from how and where they are physically stored. At the time of this writing, some use of this pattern is being replaced by the emerging Kite SDK for managing data in Hadoop. Each entry in the retrieved PCollection
Integrating Healthcare Data
There are dozens of processing steps between raw data and answers to healthcare-related questions. Here we look at one: bringing together data for a single person from multiple sources.
Unfortunately, the lack of a common patient identifier in the United States, combined with noisy data such as variations in a person’s name and demographics between systems, makes it difficult to accurately unify a person’s data across sources. Information spread across multiple sources might look like Table 22-2.
Table 22-2. Data from multiple sources
Source Person ID First name Last name Address Gender
| Doctor’s office 12345 | Abraham Lincoln | 1600 Pennsylvania Ave. M |
|---|---|---|
| Hospital 98765 | Abe Lincoln | Washington, DC M |
| Hospital 45678 | Mary Todd Lincoln | 1600 Pennsylvania Ave. F |
| Clinic 76543 | A. Lincoln | Springfield, IL M |
This is typically resolved in healthcare by a system called an Enterprise Master Patient Index (EMPI). An EMPI can be fed data from multiple systems and determine which records are indeed for the same person. This is achieved in a variety of ways, ranging from humans explicitly stating relationships to sophisticated algorithms that identify commonality.
In some cases, we can load EMPI information from external systems, and in others we compute it within Hadoop. The key is that we can expose this information for use in our Crunch-based pipelines. The result is a PCollection
@namespace(“com.cerner.example”) protocol EMPIProtocol {
record PersonRecordId { string sourceId; string personId }
/*
Represents an EMPI match.
*/
record EMPIRecord { string empiId; array
} }
Given EMPI information for the data in this structure, PCollection
Table 22-3. EMPI data
EMPI identifier PersonRecordIds (
| EMPI-1 | |
|---|---|
| EMPI-2 |
In order to group a person’s medical records in a single location based upon the provided
PCollection
The first step is to extract the common key from each EMPIRecord in the collection:
PCollection
PTable
@Override
public void process(EMPIRecord input,
Emitter
Pair.of(recordId.getSourceId(), recordId.getPersonId()), input));
}
}
}, tableOf(pairs(strings(), strings()), records(EMPIRecord.class) );
Next, the same key needs to be extracted from each PersonRecord:
PCollection
PTable
@Override
public Pair
}
}, pairs(strings(), strings()));
Joining the two PTable objects will return a PTable
Pair
PTable
.join(keyedEmpiRecords)
.values()
.by(new MapFn
@Override
public String map(Pair
}
}, strings()));
The final step is to group the table by its key to ensure all of the data is aggregated together for processing as a complete collection:
PGroupedTable
The PGroupedTable would contain data like that in Table 22-4.
This logic to unify data sources is the first step of a larger execution flow. Other Crunch functions downstream build on these steps to meet many client needs. In a common use case, a number of problems are solved by loading the contents of the unified
PersonRecords into a rules-based processing model to emit new clinical knowledge. For instance, we may run rules over those records to determine if a diabetic is receiving recommended care, and to indicate areas that can be improved. Similar rule sets exist for a variety of needs, ranging from general wellness to managing complicated conditions. The logic can be complicated and with a lot of variance between use cases, but it is all hosted in functions composed in a Crunch pipeline.
Table 22-4. Grouped EMPI data
EMPI identifier Iterable
EMPI-1 {
“personId”: “12345”,
“demographics”: {
“firstName”: “Abraham”, “lastName”: “Lincoln”, …
},
“labResults”: […]
},
{
“personId”: “98765”,
“demographics”: {
“firstName”: “Abe”, “lastName”: “Lincoln”, …
},
“diagnoses”: […]
},
{
“personId”: “98765”,
“demographics”: {
“firstName”: “Abe”, “lastName”: “Lincoln”, …
},
“medications”: […]},
{
“personId”: “76543”,
“demographics”: {
“firstName”: “A.”, “lastName”: “Lincoln”, …
} … }
| EMPI-2 | { “personId”: “45678”, “demographics”: { “firstName”: “Mary Todd”, “lastName”: “Lincoln”, … } … } |
|---|---|
Composability over Frameworks
The patterns described here take on a particular class of problem in healthcare centered around the person. However, this data can serve as the basis for understanding operational and systemic properties of healthcare as well, creating new demands on our ability to transform and analyze it.
Libraries like Crunch help us meet emerging demands because they help make our data and processing logic composable. Rather than a single, static framework for data processing, we can modularize functions and datasets and reuse them as new needs emerge. Figure 22-2 shows how components can be wired into one another in novel ways, with each box implemented as one or more Crunch DoFns. Here we leverage person records to identify diabetics and recommend health management programs, while using those composable pieces to integrate operational data and drive analytics of the health system.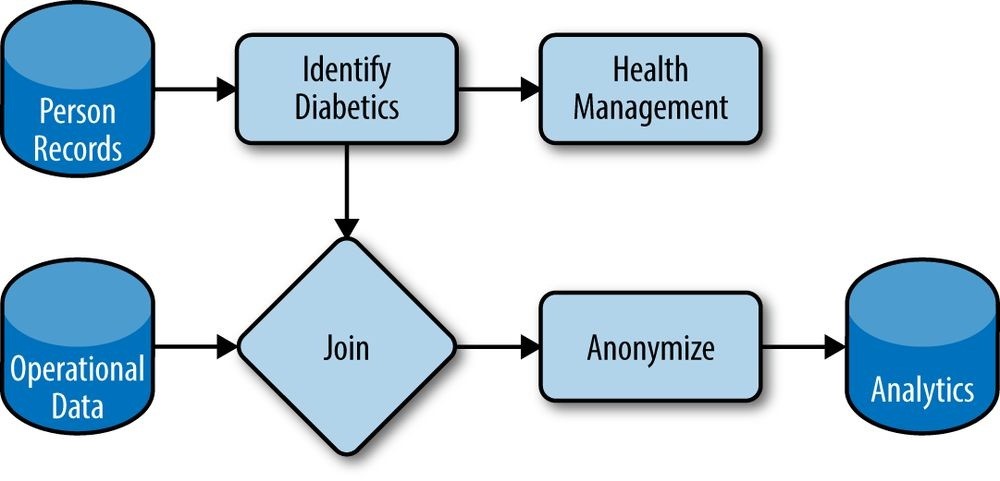
Figure 22-2. Composable datasets and functions
Composability also makes iterating through new problem spaces easier. When creating a new view of data to answer a new class of question, we can tap into existing datasets and transformations and emit our new version. As the problem becomes better understood, that view can be replaced or updated iteratively. Ultimately, these new functions and datasets can be contributed back and leveraged for new needs. The result is a growing catalog of datasets to support growing demands to understand the data.
Processing is orchestrated with Oozie. Every time new data arrives, a new dataset is created with a unique identifier in a well-defined location in HDFS. Oozie coordinators watch that location and simply launch Crunch jobs to create downstream datasets, which may subsequently be picked up by other coordinators. At the time of this writing, datasets and updates are identified by UUIDs to keep them unique. However, we are in the process of placing new data in timestamp-based partitions in order to better work with Oozie’s nominal time model.
Moving Forward
We are looking to two major steps to maximize the value from this system more efficiently.
First, we want to create prescriptive practices around the Hadoop ecosystem and its supporting libraries. A number of good practices are defined in this book and elsewhere, but they often require significant expertise to implement effectively. We are using and building libraries that make such patterns explicit and accessible to a larger audience. Crunch offers some good examples of this, with a variety of join and processing patterns built into the library.
Second, our growing catalog of datasets has created a demand for simple and prescriptive data management to complement the processing features offered by Crunch. We have been adopting the Kite SDK to meet this need in some use cases, and expect to expand its use over time.
The end goal is a secure, scalable catalog of data to support many needs in healthcare, including problems that have not yet emerged. Hadoop has shown it can scale to our data and processing needs, and higher-level libraries are now making it usable by a larger audience for many problems.
Chapter 23. Biological Data Science:
Saving Lives with Software
Matt Massie
It’s hard to believe a decade has passed since the MapReduce paper appeared at OSDI’04. It’s also hard to overstate the impact that paper had on the tech industry; the MapReduce paradigm opened distributed programming to nonexperts and enabled large-scale data processing on clusters built using commodity hardware. The open source community responded by creating open source MapReduce-based systems, like Apache Hadoop and Spark, that enabled data scientists and engineers to formulate and solve problems at a scale unimagined before.
While the tech industry was being transformed by MapReduce-based systems, biology was experiencing its own metamorphosis driven by second-generation (or “nextgeneration”) sequencing technology; see Figure 23-1. Sequencing machines are scientific instruments that read the chemical “letters” (A, C, T, and G) that make up your genome:
your complete set of genetic material. To have your genome sequenced when the MapReduce paper was published cost about $20 million and took many months to complete; today, it costs just a few thousand dollars and takes only a few days. While the first human genome took decades to create, in 2014 alone an estimated 228,000 genomes were sequenced worldwide.152] This estimate implies around 20 petabytes (PB) of sequencing data were generated in 2014 worldwide.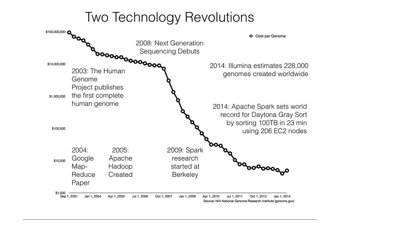
The plummeting cost of sequencing points to superlinear growth of genomics data over the coming years. This DNA data deluge has left biological data scientists struggling to process data in a timely and scalable way using current genomics software. The AMPLab is a research lab in the Computer Science Division at UC Berkeley focused on creating novel big data systems and applications. For example, Apache Spark (see Chapter 19) is one system that grew out of the AMPLab. Spark recently broke the world record for the Daytona Gray Sort, sorting 100 TB in just 23 minutes. The team at Databricks that broke the record also demonstrated they could sort 1 PB in less than 4 hours!
Consider this amazing possibility: we have technology today that could analyze every genome collected in 2014 on the order of days using a few hundred machines.
While the AMPLab identified genomics as the ideal big data application for technical reasons, there are also more important compassionate reasons: the timely processing of biological data saves lives. This short use case will focus on systems we use and have developed, with our partners and the open source community, to quickly analyze large biological datasets.
The Structure of DNA
The discovery in 1953 by Francis Crick and James D. Watson, using experimental data collected by Rosalind Franklin and Maurice Wilkins, that DNA has a double helix structure was one of the greatest scientific discoveries of the 20th century. Their Nature _article entitled “Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid” contains one of the most profound and understated sentences in science:
It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material.
This “specific pairing” referred to the observation that the bases adenine (A) and thymine
(T) always pair together and guanine (G) and cytosine (C) always pair together; see Figure 23-2. This deterministic pairing enables a “copying mechanism”: the DNA double helix unwinds and complementary base pairs snap into place, creating two exact copies of the original DNA strand.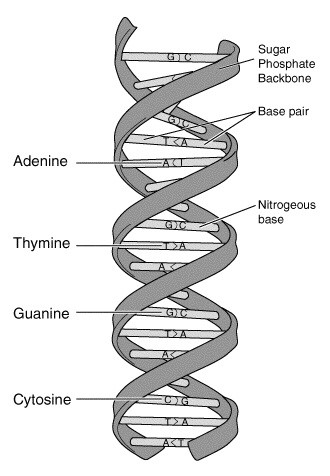
_Figure 23-2. DNA double helix structure
The Genetic Code: Turning DNA Letters into Proteins
Without proteins, there is no life. DNA serves as a recipe for creating proteins. A protein is a chain of amino acids that folds into a specific 3D shape153] to serve a particular
structure or function. As there are a total of 20 amino acids154] and only four letters in the DNA alphabet (A, C, T, G), nature groups these letters in words, called codons. Each codon is three bases long (since two bases would only support 4=16 amino acids).
In 1968, Har Gobind Khorana, Robert W. Holley, and Marshall Nirenberg received the Nobel Prize in Physiology or Medicine for successfully mapping amino acids associated with each of the 64 codons. Each codon encodes a single amino acid, or designates the start and stop positions (see Table 23-1). Since there are 64 possible codons and only 20 amino acids, multiple codons correspond to some of the amino acids.
Table 23-1. Codon table
Amino acid Codon(s) Amino acid Codon(s)
| Alanine | GC{U,C,A,G} Leucine | UU{A,G} or CU{U,C,A,G} | |
|---|---|---|---|
| Arginine | CG{U,C,A,G} or AG{A,G} Lysine | AA{A,G} | |
| Asparagine | AA{U,C} | Methionine | AUG |
| Aspartic acid | GA{U,C} | Phenylalanine | UU{U,C} |
| Cysteine | UG{U,C} | Proline | CC{U,C,A,G} |
| Glutamic acid | GA{A,G} | Threonine | AC{U,C,A,G} |
| Glutamine | CA{A,G} | Serine | UC{U,C,A,G} or AG{U,C} |
| Glycine | GG{U,C,A,G} | Tryptophan | UGG |
| Histidine | CA{U,C} | Tyrosine | UA{U,C} |
| Isoleucine | AU{U,C,A} | Valine | GU{U,C,A,G} |
| START! | AUG | STOP! | UAA or UGA or UAG |
Because every organism on Earth evolved from the same common ancestor, every organism on Earth uses the same genetic code, with few variations. Whether the organism is a tree, worm, fungus, or cheetah, the codon UGG encodes tryptophan. Mother Nature has been the ultimate practitioner of code reuse over the last few billion years.
DNA is not directly used to synthesize amino acids. Instead, a process called transcription _copies the DNA sequence that codes for a protein into _messenger RNA (mRNA). These mRNA carry information from the nuclei of your cells to the surrounding cytoplasm to create proteins in a process called translation.
You probably noticed that this lookup table doesn’t have the DNA letter T (for thymine) and has a new letter U (for uracil). During transcription, U is substituted for T:
$ echo “ATGGTGACTCCTACATGA” | sed ‘s/T/U/g’ | fold -w 3 AUG
GUG
ACU
CCU
ACA
UGA
Looking up these codons in the codon table, we can determine that this particular DNA strand will translate into a protein with the following amino acids in a chain: methionine, valine, threonine, proline, and threonine. This is a contrived example, but it logically demonstrates how DNA instructs the creation of proteins that make you uniquely you. It’s a marvel that science has allowed us to understand the language of DNA, including the start and stop punctuations.
Thinking of DNA as Source Code
At the cellular level, your body is a completely distributed system. Nothing is centralized.
It’s like a cluster of 37.2 trillion155] cells executing the same code: your DNA.
If you think of your DNA as source code, here are some things to consider: The source is comprised of only four characters: A, C, T, and G.
The source is comprised of only four characters: A, C, T, and G.
The source has two contributors, your mother and father, who contributed 3.2 billion letters each. In fact, the reference genome provided by the Genome Reference
Consortium (GRC) is nothing more than an ASCII file with 3.2 billion characters
inside.[156] The source is broken up into 25 separate files called chromosomes that each hold varying fractions of the source. The files are numbered, and tend to get smaller in size, with chromosome 1 holding ~250 million characters and chromosome 22 holding only ~50 million. There are also the X, Y, and mitochondrial chromosomes. The term chromosome basically means “colored thing,” from a time when biologists could stain them but didn’t know what they were.
The source is broken up into 25 separate files called chromosomes that each hold varying fractions of the source. The files are numbered, and tend to get smaller in size, with chromosome 1 holding ~250 million characters and chromosome 22 holding only ~50 million. There are also the X, Y, and mitochondrial chromosomes. The term chromosome basically means “colored thing,” from a time when biologists could stain them but didn’t know what they were. The source is executed on your biological machinery three letters (i.e., a codon) at a time, using the genetic code explained previously — not unlike a Turing machine that reads chemical letters instead of paper ribbon.
The source is executed on your biological machinery three letters (i.e., a codon) at a time, using the genetic code explained previously — not unlike a Turing machine that reads chemical letters instead of paper ribbon. The source has about 20,000 functions, called genes, which each create a protein when executed. The location of each gene in the source is called the locus. You can think of a gene as a specific range of contiguous base positions on a chromosome. For example, the BRCA1 gene implicated in breast cancer can be found on chromosome 17 from positions 41,196,312 to 41,277,500. A gene is like a “pointer” or “address,” whereas alleles (described momentarily) are the actual content. Everyone has the BRCA1 gene, but not everyone has alleles that put them at risk.
The source has about 20,000 functions, called genes, which each create a protein when executed. The location of each gene in the source is called the locus. You can think of a gene as a specific range of contiguous base positions on a chromosome. For example, the BRCA1 gene implicated in breast cancer can be found on chromosome 17 from positions 41,196,312 to 41,277,500. A gene is like a “pointer” or “address,” whereas alleles (described momentarily) are the actual content. Everyone has the BRCA1 gene, but not everyone has alleles that put them at risk. A haplotype is similar to an object in object-oriented programming languages that holds specific functions (genes) that are typically inherited together.
A haplotype is similar to an object in object-oriented programming languages that holds specific functions (genes) that are typically inherited together. The source has two definitions for each gene, called alleles — one from your mother and one from your father — which are found at the same position of paired chromosomes (while the cells in your body are diploid — that is, they have two alleles per gene — there are organisms that are triploid, tetraploid, etc.). Both alleles are executed and the resultant proteins interact to create a specific phenotype. For example, proteins that make or degrade eye color pigment lead to a particular phenotype, or an observable characteristic (e.g., blue eyes). If the alleles you inherit from your parents are identical, you’re homozygous for that allele; otherwise, you’re heterozygous. A single-nucleic polymorphism (SNP), pronounced “snip,” is a single-character change in the source code (e.g., from ACTGACTG to ACTTACTG).
The source has two definitions for each gene, called alleles — one from your mother and one from your father — which are found at the same position of paired chromosomes (while the cells in your body are diploid — that is, they have two alleles per gene — there are organisms that are triploid, tetraploid, etc.). Both alleles are executed and the resultant proteins interact to create a specific phenotype. For example, proteins that make or degrade eye color pigment lead to a particular phenotype, or an observable characteristic (e.g., blue eyes). If the alleles you inherit from your parents are identical, you’re homozygous for that allele; otherwise, you’re heterozygous. A single-nucleic polymorphism (SNP), pronounced “snip,” is a single-character change in the source code (e.g., from ACTGACTG to ACTTACTG).
An indel is short for insert-delete and represents an insertion or deletion from the reference genome. For example, if the reference has CCTGACTG and your sample has four characters inserted — say, CCTGCCTAACTG — then it is an indel. Only 0.5% of the source gets translated into the proteins that sustain your life. That portion of the source is called your exome. A human exome requires a few gigabytes to store in compressed binary files.
Only 0.5% of the source gets translated into the proteins that sustain your life. That portion of the source is called your exome. A human exome requires a few gigabytes to store in compressed binary files. The other 99.5% of the source is commented out and serves as word padding (introns); it is used to regulate when genes are turned on, repeat, and so on.157] A whole genome _requires a few hundred gigabytes to store in compressed binary files.
The other 99.5% of the source is commented out and serves as word padding (introns); it is used to regulate when genes are turned on, repeat, and so on.157] A whole genome _requires a few hundred gigabytes to store in compressed binary files. Every cell of your body has the same source,158] but it can be selectively commented out by _epigenetic factors like DNA methylation and histone modification, not unlike an
Every cell of your body has the same source,158] but it can be selectively commented out by _epigenetic factors like DNA methylation and histone modification, not unlike an
#ifdef statement for each cell type (e.g., #ifdef RETINA or #ifdef LIVER). These factors are responsible for making cells in your retina operate differently than cells in your liver. The process of variant calling is similar to running diff between two different DNA sources.
The process of variant calling is similar to running diff between two different DNA sources.
These analogies aren’t meant to be taken too literally, but hopefully they helped familiarize you with some genomics terminology.
The Human Genome Project and Reference Genomes
In 1953, Watson and Crick discovered the structure of DNA, and in 1965 Nirenberg, with help from his NIH colleagues, cracked the genetic code, which expressed the rules for translating DNA or mRNA into proteins. Scientists knew that there were millions of human proteins but didn’t have a complete survey of the human genome, which made it impossible to fully understand the genes responsible for protein synthesis. For example, if each protein was created by a single gene, that would imply millions of protein-coding genes in the human genome.
In 1990, the Human Genome Project set out to determine all the chemical base pairs that make up human DNA. This collaborative, international research program published the first human genome in April of 2003,159] at an estimated cost of $3.8 billion. The Human Genome Project generated an estimated $796 billion in economic impact, equating to a return on investment (ROI) of 141:1.160] The Human Genome Project found about 20,500 genes — significantly fewer than the millions you would expect with a simple 1:1 model of gene to protein, since proteins can be assembled from a combination of genes, posttranslational processes during folding, and other mechanisms.
While this first human genome took over a decade to build, once created, it made “bootstrapping” the subsequent sequencing of other genomes much easier. For the first genome, scientists were operating in the dark. They had no reference to search as a roadmap for constructing the full genome. There is no technology to date that can read a whole genome from start to finish; instead, there are many techniques that vary in the speed, accuracy, and length of DNA fragments they can read. Scientists in the Human Genome Project had to sequence the genome in pieces, with different pieces being more easily sequenced by different technologies. Once you have a complete human genome, subsequent human genomes become much easier to construct; you can use the first genome as a reference for the second. The fragments from the second genome can be pattern matched to the first, similar to having the picture on a jigsaw puzzle’s box to help inform the placement of the puzzle pieces. It helps that most coding sequences are highly conserved, and variants only occur at 1 in 1,000 loci.
Shortly after the Human Genome Project was completed, the Genome Reference Consortium (GRC), an international collection of academic and research institutes, was formed to improve the representation of reference genomes. The GRC publishes a new human reference that serves as something like a common coordinate system or map to help analyze new genomes. The latest human reference genome, released in February 2014, was named GRCh38; it replaced GRCh37, which was released five years prior.
Sequencing and Aligning DNA
Second-generation sequencing is rapidly evolving, with numerous hardware vendors and new sequencing methods being developed about every six months; however, a common feature of all these technologies is the use of massively parallel methods, where thousands or even millions of reactions occur simultaneously. The double-stranded DNA is split down the middle, the single strands are copied many times, and the copies are randomly shredded into small fragments of different lengths called reads, which are placed into the sequencer. The sequencer reads the “letters” in each of these reads, in parallel for high throughput, and outputs a raw ASCII file containing each read (e.g., AGTTTCGGGATC…), as well as a quality estimate for each letter read, to be used for downstream analysis.
A piece of software called an aligner takes each read and works to find its position in the reference genome (see Figure 23-3).161] A complete human genome is about 3 billion base (A, C, T, G) pairs long.162] The reference genome (e.g., GRCh38) acts like the picture on a puzzle box, presenting the overall contours and colors of the human genome. Each short read is like a puzzle piece that needs to be fit into position as closely as possible. A common metric is “edit distance,” which quantifies the number of operations necessary to transform one string to another. Identical strings have an edit distance of zero, and an indel of one letter has an edit distance of one. Since humans are 99.9% identical to one another, most of the reads will fit to the reference quite well and have a low edit distance. The challenge with building a good aligner is handling idiosyncratic reads.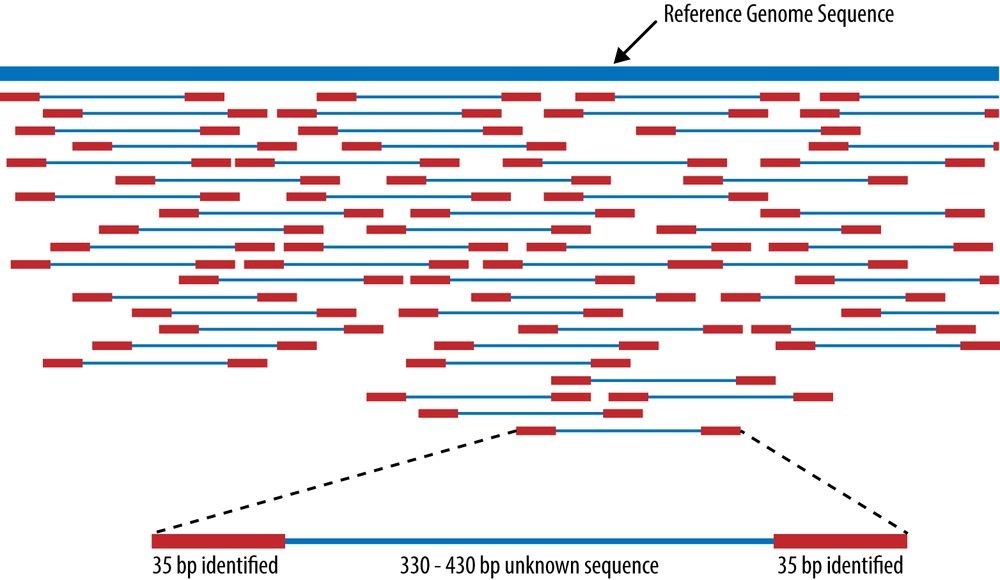
Figure 23-3. Aligning reads to a reference genome, from Wikipedia
ADAM, A Scalable Genome Analysis Platform
Aligning the reads to a reference genome is only the first of a series of steps necessary to generate reports that are useful in a clinical or research setting. The early stages of this processing pipeline look similar to any other extract-transform-load (ETL) pipelines that need data deduplication and normalization before analysis.
The sequencing process duplicates genomic DNA, so it’s possible that the same DNA reads are generated multiple times; these duplicates need to be marked. The sequencer also provides a quality estimate for each DNA “letter” that it reads, which has sequencerspecific biases that need to be adjusted. Aligners often misplace reads that have indels (inserted or deleted sequences) that need to be repositioned on the reference genome. Currently, this preprocessing is done using single-purpose tools launched by shell scripts on a single machine. These tools take multiple days to finish the processing of whole genomes. The process is disk bound, with each stage writing a new file to be read into subsequent stages, and is an ideal use case for applying general-purpose big data technology. ADAM is able to handle the same preprocessing in under two hours.
ADAM is a genome analysis platform that focuses on rapidly processing petabytes of high-coverage, whole genome data. ADAM relies on Apache Avro, Parquet, and Spark. These systems provide many benefits when used together, since they: Allow developers to focus on algorithms without needing to worry about distributed system failures
Allow developers to focus on algorithms without needing to worry about distributed system failures
Enable jobs to be run locally on a single machine, on an in-house cluster, or in the cloud without changing code
Compress legacy genomic formats and provide predicate pushdown and projection for performance
Provide an agile way of customizing and evolving data formats
Are designed to easily scale out using only commodity hardware
Are shared with a standard Apache 2.0 license163]
Literate programming with the Avro interface description language (IDL)
The Sequence Alignment/Map (SAM) specification defines the mandatory fields listed in Table 23-2.
Table 23-2. Mandatory fields in the SAM format
| Col | Field | Type | Regexp/Range | Brief description |
|---|---|---|---|---|
| 1 | QNAME | String | [!-?A-~]{1,255} | Query template NAME |
| 2 | FLAG | Int | [0, 216-1] | bitwise FLAG |
| 3 | RNAME | String | \|[!-()+-<>-~][!-~] | Reference sequence NAME |
| 4 | POS | Int | [0,231-1] | 1-based leftmost mapping POSition |
| 5 | MAPQ | Int | [0,28-1] | MAPping Quality |
| 6 | CIGAR | String | \*|([0-9]+[MIDNSHPX=])+ | CIGAR string |
| 7 | RNEXT | String | \|=|[!-()+-><-~][!-~] | Ref. name of the mate/NEXT read |
| 8 | PNEXT | Int | [0,231-1] | Position of the mate/NEXT read |
| 9 | TLEN | Int | [-231+1,231-1] | observed Template LENgth |
| 10 | SEQ | String | \*|[A-Za-z=.]+ | segment SEQuence |
| 11 | QUAL | String | [!-~] | ASCII of Phred-scaled base QUALity+33 |
Any developers who want to implement this specification need to translate this English spec into their computer language of choice. In ADAM, we have chosen instead to use literate programming with a spec defined in Avro IDL. For example, the mandatory fields for SAM can be easily expressed in a simple Avro record:
record AlignmentRecord { string qname; int flag; string rname; int pos; int mapq; string cigar; string rnext; int pnext; int tlen; string seq; string qual; }
Avro is able to autogenerate native Java (or C++, Python, etc.) classes for reading and writing data and provides standard interfaces (e.g., Hadoop’s InputFormat) to make integration with numerous systems easy. Avro is also designed to make schema evolution easier. In fact, the ADAM schemas we use today have evolved to be more sophisticated, expressive, and customized to express a variety of genomic models such as structural variants, genotypes, variant calling annotations, variant effects, and more.
UC Berkeley is a member of the Global Alliance for Genomics & Health, a non-
governmental, public-private partnership consisting of more than 220 organizations across 30 nations, with the goal of maximizing the potential of genomics medicine through effective and responsible data sharing. The Global Alliance has embraced this literate programming approach and publishes its schemas in Avro IDL as well. Using Avro has allowed researchers around the world to talk about data at the logical level, without concern for computer languages or on-disk formats. Column-oriented access with Parquet
The SAM and BAM164] file formats are row-oriented: the data for each record is stored together as a single line of text or a binary record. (See Other File Formats and ColumnOriented Formats for further discussion of row- versus column-oriented formats.) A single paired-end read in a SAM file might look like this:
read1 99 chrom1 7 30 8M2I4M1D3M = 37 39 TTAGATAAAGGATACTG read1 147 chrom1 37 30 9M = 7 -39 CAGCGGCAT NM:i:1
A typical SAM/BAM file contains many millions of rows, one for each DNA read that came off the sequencer. The preceding text fragment translates loosely into the view shown in Table 23-3.
Table 23-3. Logical view of SAM fragment
| Name | Reference | Position | MapQ | CIGAR | Sequence |
|---|---|---|---|---|---|
| read1 | chromosome1 | 7 | 30 | 8M2I4M1D3M | TTAGATAAAGGATACTG |
| read1 | chromosome1 | 37 | 30 | 9M | CAGCGGCAT |
In this example, the read, identified as read1, was mapped to the reference genome at chromosome1, positions 7 and 37. This is called a “paired-end” read as it represents a single strand of DNA that was read from each end by the sequencer. By analogy, it’s like reading an array of length 150 from 0..50 and 150..100.
The MapQ score represents the probability that the sequence is mapped to the reference correctly. MapQ scores of 20, 30, and 40 have a probability of being correct of 99%, 99.9%, and 99.99%, respectively. To calculate the probability of error from a MapQ score, use the expression 10 (e.g., 10 is a probability of 0.001).
The CIGAR explains how the individual nucleotides in the DNA sequence map to the reference.165] The Sequence is, of course, the DNA sequence that was mapped to the reference.
There is a stark mismatch between the SAM/BAM row-oriented on-disk format and the column-oriented access patterns common to genome analysis. Consider the following: A range query to find data for a particular gene linked to breast cancer, named BRCA1:
A range query to find data for a particular gene linked to breast cancer, named BRCA1:
“Find all reads that cover chromosome 17 from position 41,196,312 to 41,277,500”
A simple filter to find poorly mapped reads: “Find all reads with a MapQ less than 10” A search of all reads with insertions or deletions, called indels: “Find all reads that contain I or D in the CIGAR string”
Count the number of unique k-mers: “Read every Sequence and generate all possible substrings of length k in the string”
Parquet’s predicate pushdown feature allows us to rapidly filter reads for analysis (e.g., finding a gene, ignoring poorly mapped reads). Projection allows for precise materialization of only the columns of interest (e.g., reading only the sequences for k-mer counting).
Additionally, a number of the fields have low cardinality, making them ideal for data compression techniques like run-length encoding (RLE). For example, given that humans have only 23 pairs of chromosomes, the Reference field will have only a few dozen unique values (e.g., chromosome1, chromosome17, etc.). We have found that storing BAM records inside Parquet files results in ~20% compression. Using the PrintFooter command in Parquet, we have found that quality scores can be run-length encoded and bitpacked to compress ~48%, but they still take up ~70% of the total space. We’re looking forward to Parquet 2.0, so we can use delta encoding on the quality scores to compress the file size even more.
A simple example: k-mer counting using Spark and ADAM
Let’s do “word count” for genomics: counting k-mers. The term k-mers refers to all the possible subsequences of length k for a read. For example, if you have a read with the sequence AGATCTGAAG, the 3-mers for that sequence would be [‘AGA’, ‘GAT’, ‘ATC’, ‘TCT’, ‘CTG’, ‘TGA’, ‘GAA’, ‘AAG’]. While this is a trivial example, k-mers are useful when building structures like De Bruijn graphs for sequence assembly. In this example, we are going to generate all the possible 21-mers from our reads, count them, and then write the totals to a text file.
This example assumes that you’ve already created a SparkContext named sc. First, we create a Spark RDD of AlignmentRecords using a pushdown predicate to remove lowquality reads and a projection to only materialize the sequence field in each read:
// Load reads from ‘inputPath’ into an RDD for analysis val adamRecords: RDD[AlignmentRecord] = sc.adamLoad(args.inputPath,
// Filter out all low-quality reads that failed vendor quality checks predicate = Some(classOf[HighQualityReadsPredicate]),
// Only materialize the ‘sequence’ from each record projection = Some(Projection(AlignmentRecordField.sequence)))
Since Parquet is a column-oriented storage format, it can rapidly materialize only the sequence column and quickly skip over the unwanted fields. Next, we walk over each sequence using a sliding window of length k=21, emit a count of 1L, and then reduceByKey using the k-mer subsequence as the key to get the total counts for the input file:
// The length of k-mers we want to count val kmerLength = 21
// Process the reads into an RDD of tuples with k-mers and counts val kmers: RDD[(String, Long)] = adamRecords.flatMap(read => { read.getSequence
.toString
.sliding(kmerLength)
.map(k => (k, 1L))
}).reduceByKey { case (a, b) => a + b}
// Print the k-mers as a text file to the ‘outputPath’ _kmers.map { case (kmer, count) => s”$count,$kmer”}
.saveAsTextFile(args.outputPath)
When run on sample NA21144, chromosome 11 in the 1000 Genomes project,166] this job outputs the following:
AAAAAAAAAAAAAAAAAAAAAA, 124069
TTTTTTTTTTTTTTTTTTTTTT, 120590
ACACACACACACACACACACAC, 41528
GTGTGTGTGTGTGTGTGTGTGT, 40905
CACACACACACACACACACACA, 40795
TGTGTGTGTGTGTGTGTGTGTG, 40329
TAATCCCAGCACTTTGGGAGGC, 32122
TGTAATCCCAGCACTTTGGGAG, 31206
CTGTAATCCCAGCACTTTGGGA, 30809
GCCTCCCAAAGTGCTGGGATTA, 30716…
ADAM can do much more than just count _k-mers. Aside from the preprocessing stages already mentioned — duplicate marking, base quality score recalibration, and indel realignment — it also: Calculates coverage read depth at each variant in a Variant Call Format (VCF) file
Calculates coverage read depth at each variant in a Variant Call Format (VCF) file
Counts the k-mers/q-mers from a read dataset
Loads gene annotations from a Gene Transfer Format (GTF) file and outputs the corresponding gene models
Prints statistics on all the reads in a read dataset (e.g., % mapped to reference, number of duplicates, reads mapped cross-chromosome, etc.)
Launches legacy variant callers, pipes reads into stdin, and saves output from stdout
Comes with a basic genome browser to view reads in a web browser
However, the most important thing ADAM provides is an open, scalable platform. All artifacts are published to Maven Central (search for group ID org.bdgenomics) to make it easy for developers to benefit from the foundation ADAM provides. ADAM data is stored in Avro and Parquet, so you can also use systems like SparkSQL, Impala, Apache Pig, Apache Hive, or others to analyze the data. ADAM also supports job written in Scala, Java, and Python, with more language support on the way.
At Scala.IO in Paris in 2014, Andy Petrella and Xavier Tordoir used Spark’s MLlib kmeans with ADAM for population stratification across the 1000 Genomes dataset (population stratification is the process of assigning an individual genome to an ancestral group). They found that ADAM/Spark improved performance by a factor of 150.
From Personalized Ads to Personalized Medicine
While ADAM is designed to rapidly and scalably analyze aligned reads, it does not align the reads itself; instead, ADAM relies on standard short-reads aligners. The Scalable Nucleotide Alignment Program (SNAP) is a collaborative effort including participants from Microsoft Research, UC San Francisco, and the AMPLab as well as open source developers, shared with an Apache 2.0 license. The SNAP aligner is as accurate as the current best-of-class aligners, like BWA-mem, Bowtie2, and Novalign, but runs between 3 and 20 times faster. This speed advantage is important when doctors are racing to identify a pathogen.
In 2013, a boy went to the University of Wisconsin Hospital and Clinics’ Emergency Department three times in four months with symptoms of encephalitis: fevers and headaches. He was eventually hospitalized without a successful diagnosis after numerous blood tests, brain scans, and biopsies. Five weeks later, he began having seizures that required he be placed into a medically induced coma. In desperation, doctors sampled his spinal fluid and sent it to an experimental program led by Charles Chiu at UC San Francisco, where it was sequenced for analysis. The speed and accuracy of SNAP allowed UCSF to quickly filter out all human DNA and, from the remaining 0.02% of the reads, identify a rare infectious bacterium, Leptospira santarosai. They reported the discovery to the Wisconsin doctors just two days after they sent the sample. The boy was treated with antibiotics for 10 days, awoke from his coma, and was discharged from the hospital two weeks later.167]
If you’re interested in learning more about the system the Chiu lab used — called Sequence-based Ultra-Rapid Pathogen Identification (SURPI) — they have generously shared their software with a permissive BSD license and provide an Amazon EC2 Machine Image (AMI) with SURPI preinstalled. SURPI collects 348,922 unique bacterial sequences and 1,193,607 unique virus sequences from numerous sources and saves them in 29 SNAP-indexed databases, each approximately 27 GB in size, for fast search.
Today, more data is analyzed for personalized advertising than personalized medicine, but that will not be the case in the future. With personalized medicine, people receive customized healthcare that takes into consideration their unique DNA profiles. As the price of sequencing drops and more people have their genomes sequenced, the increase in statistical power will allow researchers to understand the genetic mechanisms underlying diseases and fold these discoveries into the personalized medical model, to improve treatment for subsequent patients. While only 25 PB of genomic data were generated worldwide this year, next year that number will likely be 100 PB.
Join In
While we’re off to a great start, the ADAM project is still an experimental platform and needs further development. If you’re interested in learning more about programming on ADAM or want to contribute code, take a look at Advanced Analytics with Spark: Patterns for Learning from Data at Scale by Sandy Ryza et al. (O’Reilly, 2014), which includes a chapter on analyzing genomics data with ADAM and Spark. You can find us at http://bdgenomics.org, on IRC at #adamdev, or on Twitter at @bigdatagenomics.
[152] See Antonio Regalado, “EmTech: Illumina Says 228,000 Human Genomes Will Be Sequenced This Year,” September 24, 2014.
[153] This process is called protein folding. The Folding@home allows volunteers to donate CPU cycles to help researchers determine the mechanisms of protein folding.
[154] There are also a few nonstandard amino acids not shown in the table that are encoded differently.
[155] See Eva Bianconi et al., “An estimation of the number of cells in the human body,” Annals of Human Biology, November/December 2013.
[156] You might expect this to be 6.4 billion letters, but the reference genome is, for better or worse, a haploid _representation of the average of dozens of individuals.
[157] Only about 28% of your DNA is transcribed into nascent RNA, and after RNA splicing, only about 1.5% of the RNA is left to code for proteins. Evolutionary selection occurs at the DNA level, with most of your DNA providing support to the other 0.5% or being deselected altogether (as more fitting DNA evolves). There are some cancers that appear to be caused by dormant regions of DNA being resurrected, so to speak.
[158] There is actually, on average, about 1 error for each billion DNA “letters” copied. So, each cell isn’t _exactly the same.
[159] Intentionally 50 years after Watson and Crick’s discovery of the 3D structure of DNA.
[160] Jonathan Max Gitlin, “Calculating the economic impact of the Human Genome Project,” June 2013.
[161] There is also a second approach, de novo assembly, where reads are put into a graph data structure to create long sequences without mapping to a reference genome.
[162] Each base is about 3.4 angstroms, so the DNA from a single human cell stretches over 2 meters end to end!
[163] Unfortunately, some of the more popular software in genomics has an ill-defined or custom, restrictive license. Clean open source licensing and source code are necessary for science to make it easier to reproduce and understand results.
[164] BAM is the compressed binary version of the SAM format.
[165] The first record’s Compact Idiosyncratic Gap Alignment Report (CIGAR) string is translated as “8 matches (8M), 2 inserts (2I), 4 matches (4M), 1 delete (1D), 3 matches (3M).”
[166] Arguably the most popular publicly available dataset, found at http://www.1000genomes.org.
[167] Michael Wilson et al., “Actionable Diagnosis of Neuroleptospirosis by Next-Generation Sequencing,” New England Journal of Medicine, June 2014.
Chapter 24. Cascading
Chris K. Wensel
Cascading is an open source Java library and API that provides an abstraction layer for MapReduce. It allows developers to build complex, mission-critical data processing applications that run on Hadoop clusters.
The Cascading project began in the summer of 2007. Its first public release, version 0.1, launched in January 2008. Version 1.0 was released in January 2009. Binaries, source code, and add-on modules can be downloaded from the project website.
Map and reduce operations offer powerful primitives. However, they tend to be at the wrong level of granularity for creating sophisticated, highly composable code that can be shared among different developers. Moreover, many developers find it difficult to “think” in terms of MapReduce when faced with real-world problems.
To address the first issue, Cascading substitutes the keys and values used in MapReduce with simple field names and a data tuple model, where a tuple is simply a list of values. For the second issue, Cascading departs from map and reduce operations directly by introducing higher-level abstractions as alternatives: Functions, Filters, Aggregators, and Buffers.
Other alternatives began to emerge at about the same time as the project’s initial public release, but Cascading was designed to complement them. Consider that most of these alternative frameworks impose pre- and post-conditions, or other expectations.
For example, in several other MapReduce tools, you must preformat, filter, or import your data into HDFS prior to running the application. That step of preparing the data must be performed outside of the programming abstraction. In contrast, Cascading provides the means to prepare and manage your data as integral parts of the programming abstraction.
This case study begins with an introduction to the main concepts of Cascading, then finishes with an overview of how ShareThis uses Cascading in its infrastructure.
See the Cascading User Guide on the project website for a more in-depth presentation of the Cascading processing model.
Fields, Tuples, and Pipes
The MapReduce model uses keys and values to link input data to the map function, the map function to the reduce function, and the reduce function to the output data.
But as we know, real-world Hadoop applications usually consist of more than one MapReduce job chained together. Consider the canonical word count example implemented in MapReduce. If you needed to sort the numeric counts in descending order, which is not an unlikely requirement, it would need to be done in a second MapReduce job.
So, in the abstract, keys and values not only bind map to reduce, but reduce to the next map, and then to the next reduce, and so on (Figure 24-1). That is, key-value pairs are sourced from input files and stream through chains of map and reduce operations, and finally rest in an output file. When you implement enough of these chained MapReduce applications, you start to see a well-defined set of key-value manipulations used over and over again to modify the key-value data stream.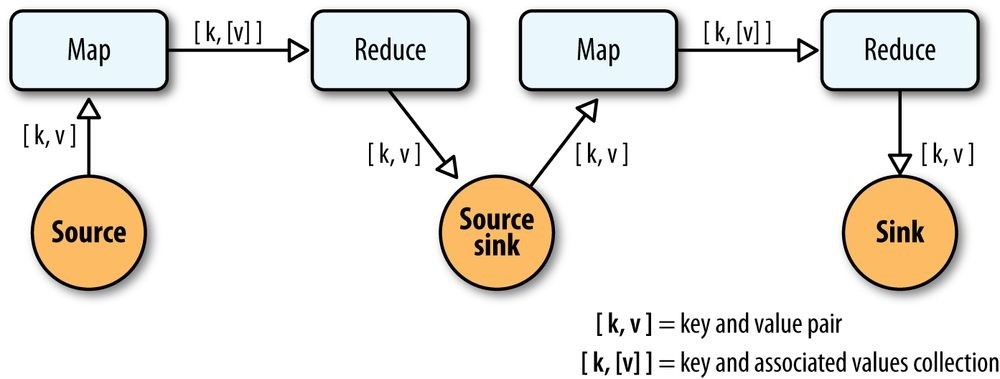
Figure 24-1. Counting and sorting in MapReduce
Cascading simplifies this by abstracting away keys and values and replacing them with tuples that have corresponding field names, similar in concept to tables and column names in a relational database. During processing, streams of these fields and tuples are then manipulated as they pass through user-defined operations linked together by pipes (Figure 24-2).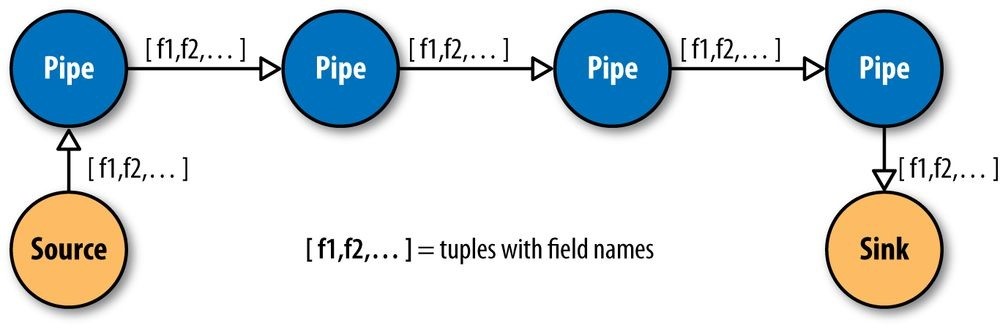
Figure 24-2. Pipes linked by fields and tuples
So, MapReduce keys and values are reduced to:
Fields
A field is a collection of either String names (such as “firstname”), numeric positions (such as 2 or –1, for the third and last positions, respectively), or a combination of both. So, fields are used to declare the names of values in a tuple and to select values by name from a tuple. The latter is like a SQL select call.
Tuples
A tuple is simply an array of java.lang.Comparable objects. A tuple is very much like a database row or record.
And the map and reduce operations are abstracted behind one or more pipe instances (Figure 24-3):
Each
The Each pipe processes a single input tuple at a time. It may apply either a Function or a Filter operation (described shortly) to the input tuple.
GroupBy
The GroupBy pipe groups tuples on grouping fields. It behaves just like the SQL GROUP BY statement. It can also merge multiple input tuple streams into a single stream if they all share the same field names.
CoGroup
The CoGroup pipe joins multiple tuple streams together by common field names, and it also groups the tuples by the common grouping fields. All standard join types (inner, outer, etc.) and custom joins can be used across two or more tuple streams.
Every
The Every pipe processes a single grouping of tuples at a time, where the group was grouped by a GroupBy or CoGroup pipe. The Every pipe may apply either an Aggregator or a Buffer operation to the grouping.
SubAssembly
The SubAssembly pipe allows for nesting of assemblies inside a single pipe, which can, in turn, be nested in more complex assemblies.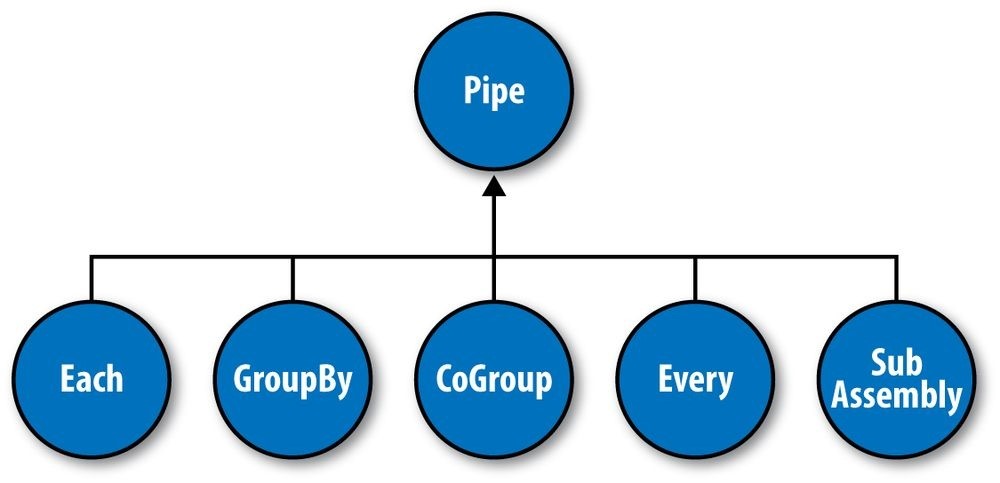
_Figure 24-3. Pipe types
All these pipes are chained together by the developer into “pipe assemblies,” in which each assembly can have many input tuple streams (sources) and many output tuple streams (sinks). See Figure 24-4.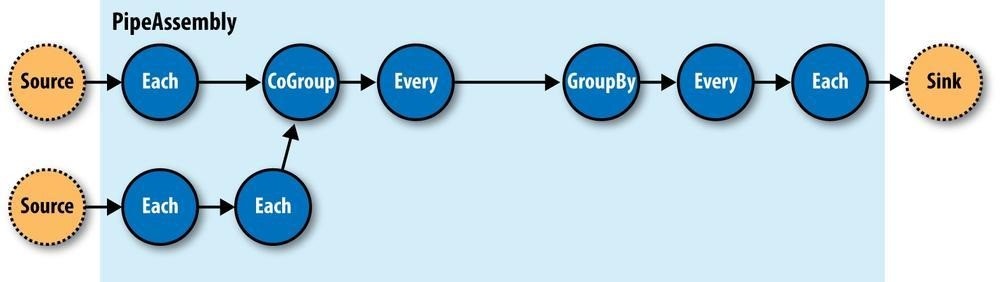
Figure 24-4. A simple PipeAssembly
On the surface, this might seem more complex than the traditional MapReduce model. And admittedly, there are more concepts here than map, reduce, key, and value. But in practice, there are many more concepts that must all work in tandem to provide different behaviors.
For example, a developer who wanted to provide a “secondary sorting” of reducer values would need to implement a map, a reduce, a “composite” key (two keys nested in a parent key), a value, a partitioner, an “output value grouping” comparator, and an “output key” comparator, all of which would be coupled to one another in varying ways, and very likely would not be reusable in subsequent applications.
In Cascading, this would be one line of code: new GroupBy(
Operations
As mentioned earlier, Cascading departs from MapReduce by introducing alternative operations that are applied either to individual tuples or groups of tuples (Figure 24-5):
Function
A Function operates on individual input tuples and may return zero or more output tuples for every one input. Functions are applied by the Each pipe.
Filter
A Filter is a special kind of function that returns a Boolean value indicating whether the current input tuple should be removed from the tuple stream. A Function could serve this purpose, but the Filter is optimized for this case, and many filters can be grouped by “logical” filters such as AND, OR, XOR, and NOT, rapidly creating more complex filtering operations.
Aggregator
An Aggregator performs some operation against a group of tuples, where the grouped tuples are by a common set of field values (for example, all tuples having the same “last-name” value). Common Aggregator implementations would be Sum, Count, Average, Max, and Min.
Buffer
A Buffer is similar to an Aggregator, except it is optimized to act as a “sliding window” across all the tuples in a unique grouping. This is useful when the developer needs to efficiently insert missing values in an ordered set of tuples (such as a missing date or duration) or create a running average. Usually Aggregator is the operation of choice when working with groups of tuples, since many Aggregators can be chained together very efficiently, but sometimes a Buffer is the best tool for the job.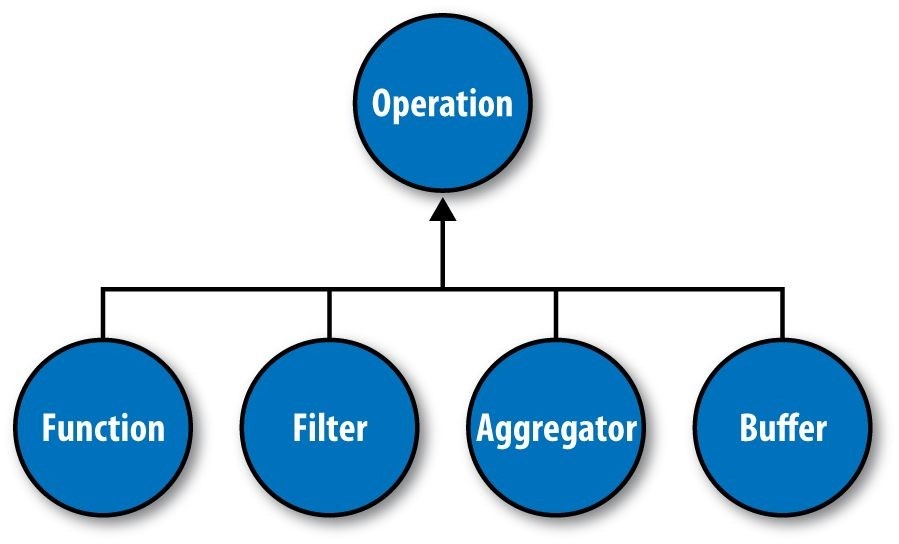
Figure 24-5. Operation types
Operations are bound to pipes when the pipe assembly is created (Figure 24-6).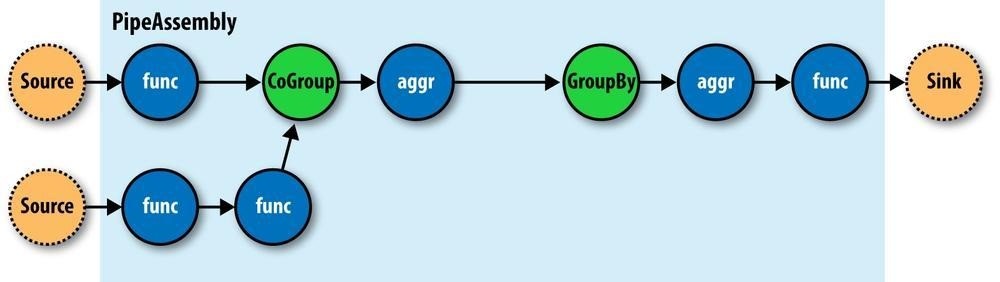
Figure 24-6. An assembly of operations
The Each and Every pipes provide a simple mechanism for selecting some or all values out of an input tuple before the values are passed to its child operation. And there is a simple mechanism for merging the operation results with the original input tuple to create the output tuple. Without going into great detail, this allows for each operation to care only about argument tuple values and fields, not the whole set of fields in the current input tuple. Subsequently, operations can be reusable across applications in the same way that Java methods can be reusable.
For example, in Java, a method declared as concatenate(String first, String second) is more abstract than concatenate(Person person). In the second case, the concatenate() function must “know” about the Person object; in the first case, it is agnostic to where the data came from. Cascading operations exhibit this same quality.
Taps, Schemes, and Flows
In many of the previous diagrams, there are references to “sources” and “sinks.” In Cascading, all data is read from or written to Tap instances, but is converted to and from tuple instances via Scheme objects:
Tap
A Tap is responsible for the “how” and “where” parts of accessing data. For example, is the data on HDFS or the local filesystem? In Amazon S3 or over HTTP?
Scheme
A Scheme is responsible for reading raw data and converting it to a tuple and/or writing a tuple out into raw data, where this “raw” data can be lines of text, Hadoop binary sequence files, or some proprietary format.
Note that Taps are not part of a pipe assembly, and so they are not a type of Pipe. But they are connected with pipe assemblies when they are made cluster executable. When a pipe assembly is connected with the necessary number of source and sink Tap instances, we get a Flow. The Taps either emit or capture the field names the pipe assembly expects. That is, if a Tap emits a tuple with the field name “line” (by reading data from a file on HDFS), the head of the pipe assembly must be expecting a “line” value as well. Otherwise, the process that connects the pipe assembly with the Taps will immediately fail with an error.
So pipe assemblies are really data process definitions, and are not “executable” on their own. They must be connected to source and sink Tap instances before they can run on a cluster. This separation between Taps and pipe assemblies is part of what makes Cascading so powerful.
If you think of a pipe assembly like a Java class, then a Flow is like a Java object instance
(Figure 24-7). That is, the same pipe assembly can be “instantiated” many times into new Flows, in the same application, without fear of any interference between them. This allows pipe assemblies to be created and shared like standard Java libraries.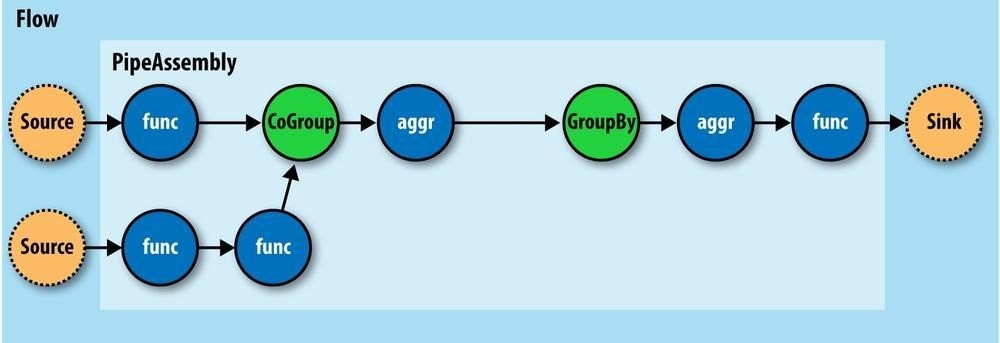
Figure 24-7. A Flow
Cascading in Practice
Now that we know what Cascading is and have a good idea of how it works, what does an application written in Cascading look like? See Example 24-1.
Example 24-1. Word count and sort
Scheme sourceScheme =
new TextLine(new Fields(“line”));  Tap source =
Tap source =
new Hfs(sourceScheme, inputPath); 
Scheme sinkScheme = new TextLine();  Tap sink =
Tap sink =
new Hfs(sinkScheme, outputPath, SinkMode.REPLACE); 
Pipe assembly = new Pipe(“wordcount”); 
String regexString = “(?<!\pL)(?=\pL)[^ ](?<=\pL)(?!\pL)”;
Function regex = new RegexGenerator(new Fields(“word”), regexString); assembly = new Each(assembly, new Fields(“line”), regex); 
assembly =
new GroupBy(assembly, new Fields(“word”)); 
Aggregator count = new Count(new Fields(“count”)); assembly = new Every(assembly, count); 
assembly =
new GroupBy(assembly, new Fields(“count”), new Fields(“word”)); 
FlowConnector flowConnector = *new FlowConnector(); Flow flow = flowConnector.connect(“word-count”, source, sink, assembly);  flow.complete();
flow.complete();
We create a new Scheme that reads simple text files and emits a new Tuple for each line in a field named “line,” as declared by the Fields instance.
We create a new Scheme that writes simple text files and expects a Tuple with any number of fields/values. If there is more than one value, they will be tab-delimited in the output file.
We create source and sink Tap instances that reference the input file and output directory, respectively. The sink Tap will overwrite any file that may already exist.
We construct the head of our pipe assembly and name it “wordcount.” This name is used to bind the source and sink Taps to the assembly. Multiple heads or tails would require unique names.
We construct an Each pipe with a function that will parse the “line” field into a new Tuple for each word encountered.
We construct a GroupBy pipe that will create a new Tuple grouping for each unique value in the field “word.”
We construct an Every pipe with an Aggregator that will count the number of Tuples in every unique word group. The result is stored in a field named “count.”
We construct a GroupBy pipe that will create a new Tuple grouping for each unique value in the field “count” and secondary sort each value in the field “word.” The result will be a list of “count” and “word” values with “count” sorted in increasing order.
We connect the pipe assembly to its sources and sinks in a Flow, and then execute the Flow on the cluster.
In the example, we count the words encountered in the input document, and we sort the counts in their natural order (ascending). If some words have the same “count” value, these words are sorted in their natural order (alphabetical).
One obvious problem with this example is that some words might have uppercase letters in some instances — for example, “the” and “The” when the word comes at the beginning of a sentence. We might consider inserting a new operation to force all the words to lowercase, but we realize that all future applications that need to parse words from documents should have the same behavior, so we’ll instead create a reusable pipe called SubAssembly, just like we would by creating a subroutine in a traditional application (see Example 24-2).
Example 24-2. Creating a SubAssembly
public class ParseWordsAssembly extends SubAssembly 
{
public ParseWordsAssembly(Pipe previous)
{
String regexString = “(?<!\pL)(?=\pL)[^ ](?<=\pL)(?!\pL)”; Function regex = new RegexGenerator(new Fields(“word”), regexString); previous = new Each(previous, new Fields(“line”), regex);
String exprString = “word.toLowerCase()”; Function expression =
new ExpressionFunction(new Fields(“word”), exprString, String.class);  previous = new Each(previous, *new Fields(“word”), expression);
previous = new Each(previous, *new Fields(“word”), expression);
setTails(previous); 
}
}
We subclass the SubAssembly class, which is itself a kind of Pipe.
We create a Java expression function that will call toLowerCase() on the String value in the field named “word.” We must also pass in the Java type the expression expects “word” to be — in this case, String. (Janino is used under the covers.)
We tell the SubAssembly superclass where the tail ends of our pipe subassembly are.
First, we create a SubAssembly pipe to hold our “parse words” pipe assembly. Because this is a Java class, it can be reused in any other application, as long as there is an incoming field named “word” (Example 24-3). Note that there are ways to make this function even more generic, but they are covered in the Cascading User Guide.
Example 24-3. Extending word count and sort with a SubAssembly
Scheme sourceScheme = new TextLine(new Fields(“line”));
Tap source = new Hfs(sourceScheme, inputPath);
Scheme sinkScheme = new TextLine(new Fields(“word”, “count”));
Tap sink = new Hfs(sinkScheme, outputPath, SinkMode.REPLACE); Pipe assembly = new Pipe(“wordcount”);
assembly = new ParseWordsAssembly(assembly);  assembly = new GroupBy(assembly, new Fields(“word”));
assembly = new GroupBy(assembly, new Fields(“word”));
Aggregator count = new Count(new Fields(“count”)); assembly = new Every(assembly, count); assembly = new GroupBy(assembly, new Fields(“count”), new Fields(“word”));
FlowConnector flowConnector = new FlowConnector();
Flow flow = flowConnector.connect(“word-count”, source, sink, assembly); flow.complete();
We replace Each from the previous example with our ParseWordsAssembly pipe.
Finally, we just substitute in our new SubAssembly right where the previous Every and word parser function were used in the previous example. This nesting can continue as deep as necessary.
Flexibility
Let’s take a step back and see what this new model has given us — or better yet, what it has taken away.
You see, we no longer think in terms of MapReduce jobs, or Mapper and Reducer interface implementations and how to bind or link subsequent MapReduce jobs to the ones that precede them. During runtime, the Cascading “planner” figures out the optimal way to partition the pipe assembly into MapReduce jobs and manages the linkages between them (Figure 24-8).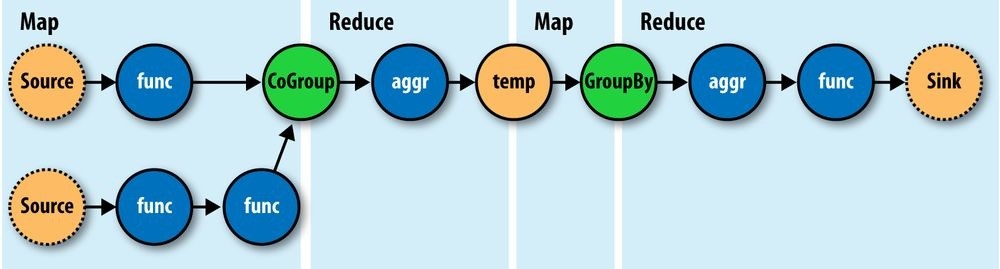
Figure 24-8. How a Flow translates to chained MapReduce jobs
Because of this, developers can build applications of arbitrary granularity. They can start with a small application that just filters a logfile, then iteratively build more features into the application as needed.
Since Cascading is an API and not a syntax like strings of SQL, it is more flexible. First off, developers can create domain-specific languages (DSLs) using their favorite languages, such as Groovy, JRuby, Jython, Scala, and others (see the project site for examples). Second, developers can extend various parts of Cascading, such as allowing custom Thrift or JSON objects to be read and written to and allowing them to be passed through the tuple stream.
Hadoop and Cascading at ShareThis
ShareThis is a sharing network that makes it simple to share any online content. With the click of a button on a web page or browser plug-in, ShareThis allows users to seamlessly access their contacts and networks from anywhere online and share the content via email, IM, Facebook, Digg, mobile SMS, and similar services, without ever leaving the current page. Publishers can deploy the ShareThis button to tap into the service’s universal sharing capabilities to drive traffic, stimulate viral activity, and track the sharing of online content. ShareThis also simplifies social media services by reducing clutter on web pages and providing instant distribution of content across social networks, affiliate groups, and communities.
As ShareThis users share pages and information through the online widgets, a continuous stream of events enter the ShareThis network. These events are first filtered and processed, and then handed to various backend systems, including AsterData, Hypertable, and Katta.
The volume of these events can be huge; too large to process with traditional systems. This data can also be very “dirty” thanks to “injection attacks” from rogue systems, browser bugs, or faulty widgets. For this reason, the developers at ShareThis chose to deploy Hadoop as the preprocessing and orchestration frontend to their backend systems. They also chose to use Amazon Web Services to host their servers on the Elastic Computing Cloud (EC2) and provide long-term storage on the Simple Storage Service (S3), with an eye toward leveraging Elastic MapReduce (EMR).
In this overview, we will focus on the “log processing pipeline” (Figure 24-9). This pipeline simply takes data stored in an S3 bucket, processes it (as described shortly), and stores the results back into another bucket. The Simple Queue Service (SQS) is used to coordinate the events that mark the start and completion of data processing runs.
Downstream, other processes pull data to load into AsterData, pull URL lists from Hypertable to source a web crawl, or pull crawled page data to create Lucene indexes for use by Katta. Note that Hadoop is central to the ShareThis architecture. It is used to coordinate the processing and movement of data between architectural components.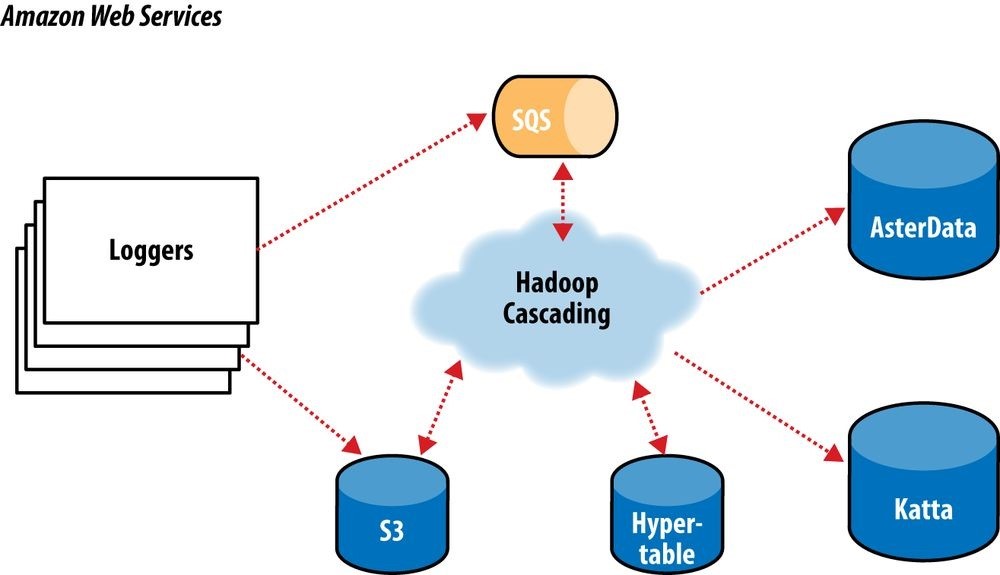
Figure 24-9. The ShareThis log processing pipeline
With Hadoop as the frontend, all the event logs can be parsed, filtered, cleaned, and organized by a set of rules before ever being loaded into the AsterData cluster or used by any other component. AsterData is a clustered data warehouse that can support large datasets and that allows for complex ad hoc queries using a standard SQL syntax. ShareThis chose to clean and prepare the incoming datasets on the Hadoop cluster and then to load that data into the AsterData cluster for ad hoc analysis and reporting. Though that process would have been possible with AsterData, it made a lot of sense to use Hadoop as the first stage in the processing pipeline to offset load on the main data warehouse.
Cascading was chosen as the primary data processing API to simplify the development process, codify how data is coordinated between architectural components, and provide the developer-facing interface to those components. This represents a departure from more “traditional” Hadoop use cases, which essentially just query stored data. Cascading and Hadoop together provide a better and simpler structure for the complete solution, end to end, and thus provide more value to the users.
For the developers, Cascading made it easy to start with a simple unit test (created by subclassing cascading.ClusterTestCase) that did simple text parsing and then to layer in more processing rules while keeping the application logically organized for maintenance.
Cascading aided this organization in a couple of ways. First, standalone operations
(Functions, Filters, etc.) could be written and tested independently. Second, the application was segmented into stages: one for parsing, one for rules, and a final stage for binning/collating the data, all via the SubAssembly base class described earlier.
The data coming from the ShareThis loggers looks a lot like Apache logs, with date/timestamps, share URLs, referrer URLs, and a bit of metadata. To use the data for analysis downstream, the URLs needed to be unpacked (parsing query-string data, domain names, etc.). So, a top-level SubAssembly was created to encapsulate the parsing, and child subassemblies were nested inside to handle specific fields if they were sufficiently complex to parse.
The same was done for applying rules. As every Tuple passed through the rules
SubAssembly, it was marked as “bad” if any of the rules were triggered. Along with the “bad” tag, a description of why the record was bad was added to the Tuple for later review.
Finally, a splitter SubAssembly was created to do two things. First, it allowed for the tuple stream to split into two: one stream for “good” data and one for “bad” data. Second, the splitter binned the data into intervals, such as every hour. To do this, only two operations were necessary: the first to create the interval from the timestamp value already present in the stream, and the second to use the interval and good/bad metadata to create a directory path (for example, 05/good/, where “05” is 5 a.m. and “good” means the Tuple passed all the rules). This path would then be used by the Cascading TemplateTap, a special Tap that can dynamically output tuple streams to different locations based on values in the Tuple. In this case, the TemplateTap used the “path” value to create the final output path.
The developers also created a fourth SubAssembly — this one to apply Cascading Assertions during unit testing. These assertions double-checked that rules and parsing subassemblies did their job.
In the unit test in Example 24-4, we see the splitter isn’t being tested, but it is added in another integration test not shown. Example 24-4. Unit testing a Flow
public void testLogParsing() throws IOException
{
Hfs source = new Hfs(new TextLine(new Fields(“line”)), sampleData); Hfs sink = new Hfs(new TextLine(), outputPath + “/parser”, SinkMode.REPLACE);
Pipe pipe = new Pipe(“parser”);
// split “line” on tabs
pipe = new Each(pipe, new Fields(“line”), new RegexSplitter(“\t”)); pipe = new LogParser(pipe);
pipe = new LogRules(pipe); // testing only assertions pipe = new ParserAssertions(pipe);
Flow flow = new FlowConnector().connect(source, sink, pipe); flow.complete(); // run the test flow
// Verify there are 98 tuples and 2 fields, and matches the regex pattern
// For TextLine schemes the tuples are { “offset”, “line” }
validateLength(flow, 98, 2, Pattern.compile(“^[0-9]+(\t[^\t]*){19}$”)); }
For integration and deployment, many of the features built into Cascading allowed for easier integration with external systems and for greater process tolerance.
In production, all the subassemblies are joined and planned into a Flow, but instead of just source and sink Taps, trap Taps were planned in (Figure 24-10). Normally, when an operation throws an exception from a remote mapper or reducer task, the Flow will fail and kill all its managed MapReduce jobs. When a Flow has traps, any exceptions are caught and the data causing the exception is saved to the Tap associated with the current trap. Then the next Tuple is processed without stopping the Flow. Sometimes you want your Flows to fail on errors, but in this case, the ShareThis developers knew they could go back and look at the “failed” data and update their unit tests while the production system kept running. Losing a few hours of processing time was worse than losing a couple of bad records.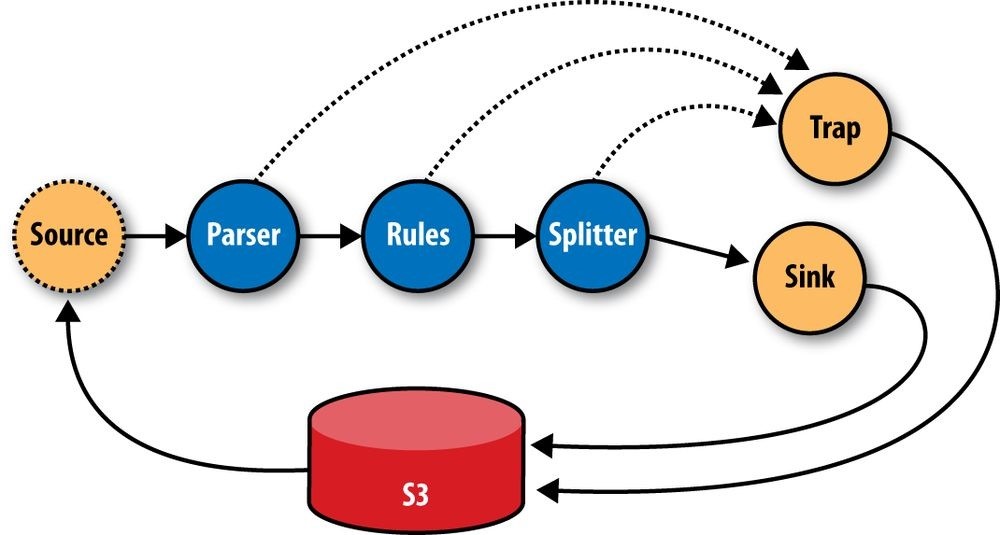
Figure 24-10. The ShareThis log processing flow
Using Cascading’s event listeners, Amazon SQS could be integrated. When a Flow finishes, a message is sent to notify other systems that there is data ready to be picked up from Amazon S3. On failure, a different message is sent, alerting other processes.
The remaining downstream processes pick up where the log processing pipeline leaves off on different independent clusters. The log processing pipeline today runs once a day; there is no need to keep a 100-node cluster sitting around for the 23 hours it has nothing to do, so it is decommissioned and recommissioned 24 hours later.
In the future, it would be trivial to increase this interval on smaller clusters to every 6 hours, or 1 hour, as the business demands. Independently, other clusters are booting and shutting down at different intervals based on the needs of the business units responsible for those components. For example, the web crawler component (using Bixo, a Cascadingbased web-crawler toolkit developed by EMI and ShareThis) may run continuously on a small cluster with a companion Hypertable cluster. This on-demand model works very well with Hadoop, where each cluster can be tuned for the kind of workload it is expected to handle.
Summary
Hadoop is a very powerful platform for processing and coordinating the movement of data across various architectural components. Its only drawback is that the primary computing model is MapReduce.
Cascading aims to help developers build powerful applications quickly and simply, through a well-reasoned API, without needing to think in MapReduce and while leaving the heavy lifting of data distribution, replication, distributed process management, and liveness to Hadoop.
Read more about Cascading, join the online community, and download sample applications by visiting the project website.
Appendix A. Installing Apache Hadoop
It’s easy to install Hadoop on a single machine to try it out. (For installation on a cluster, refer to Chapter 10.)
In this appendix, we cover how to install Hadoop Common, HDFS, MapReduce, and YARN using a binary tarball release from the Apache Software Foundation. Instructions for installing the other projects covered in this book are included at the start of the relevant chapters.
The instructions that follow are suitable for Unix-based systems, including Mac OS X (which is not a production platform, but is fine for development).
Prerequisites
Make sure you have a suitable version of Java installed. You can check the Hadoop wiki to find which version you need. The following command confirms that Java was installed correctly:
% java -version java version “1.7.0_25”
Java(TM) SE Runtime Environment (build 1.7.0_25-b15)
Java HotSpot(TM) 64-Bit Server VM (build 23.25-b01, mixed mode)
Installation
Start by deciding which user you’d like to run Hadoop as. For trying out Hadoop or developing Hadoop programs, you can run Hadoop on a single machine using your own user account.
Download a stable release, which is packaged as a gzipped tar file, from the Apache Hadoop releases page, and unpack it somewhere on your filesystem:
% tar xzf hadoop-x.y.z.tar.gz
Before you can run Hadoop, you need to tell it where Java is located on your system. If you have the JAVAHOME environment variable set to point to a suitable Java installation, that will be used, and you don’t have to configure anything further. (It is often set in a shell startup file, such as ~/.bashprofile or ~/.bashrc.) Otherwise, you can set the Java installation that Hadoop uses by editing conf/hadoop-env.sh and specifying the JAVAHOME variable. For example, on my Mac, I changed the line to read: export JAVA_HOME=/Library/Java/JavaVirtualMachines/jdk1.7.0_25.jdk/Contents/Home
to point to the installed version of Java.
It’s very convenient to create an environment variable that points to the Hadoop installation directory (HADOOP_HOME, by convention) and to put the Hadoop binary directories on your command-line path. For example:
% **export HADOOP_HOME=~/sw/hadoop-_x.y.z
% export PATH=$PATH:$HADOOPHOME/bin:$HADOOP_HOME/sbin**
Note that the _sbin directory contains the scripts for running Hadoop daemons, so it should be included if you plan to run the daemons on your local machine.
Check that Hadoop runs by typing:
% hadoop version Hadoop 2.5.1
Subversion https://git-wip-us.apache.org/repos/asf/hadoop.git -r 2e18d179e4a8065 b6a9f29cf2de9451891265cce
Compiled by jenkins on 2014-09-05T23:11Z
Compiled with protoc 2.5.0
From source with checksum 6424fcab95bfff8337780a181ad7c78
This command was run using /Users/tom/sw/hadoop-2.5.1/share/hadoop/common/hadoop -common-2.5.1.jar
Configuration
Each component in Hadoop is configured using an XML file. Common properties go in core-site.xml, and properties pertaining to HDFS, MapReduce, and YARN go into the appropriately named file: hdfs-site.xml, mapred-site.xml, and yarn-site.xml. These files are all located in the etc/hadoop subdirectory.
NOTE You can see the default settings for all the properties that are governed by these configuration files by looking in the share/doc directory hierarchy of your Hadoop installation for files called core-default.xml, hdfs-default.xml, mapreddefault.xml, and yarn-default.xml.
You can see the default settings for all the properties that are governed by these configuration files by looking in the share/doc directory hierarchy of your Hadoop installation for files called core-default.xml, hdfs-default.xml, mapreddefault.xml, and yarn-default.xml.
Hadoop can be run in one of three modes:
Standalone (or local) mode
There are no daemons running and everything runs in a single JVM. Standalone mode is suitable for running MapReduce programs during development, since it is easy to test and debug them.
Pseudodistributed mode
The Hadoop daemons run on the local machine, thus simulating a cluster on a small scale.
Fully distributed mode
The Hadoop daemons run on a cluster of machines. This setup is described in Chapter 10.
To run Hadoop in a particular mode, you need to do two things: set the appropriate properties, and start the Hadoop daemons. Table A-1 shows the minimal set of properties to configure each mode. In standalone mode, the local filesystem and the local
MapReduce job runner are used. In the distributed modes, the HDFS and YARN daemons are started, and MapReduce is configured to use YARN.
Table A-1. Key configuration properties for different modes
| Component | Property | Standalone | Pseudodistributed | Fully distributed |
|---|---|---|---|---|
| Common | fs.defaultFS | file:/// (default) |
hdfs://localhost/ | hdfs://namenode/ |
| HDFS | dfs.replication | N/A | 1 | 3 (default) |
| MapReduce | mapreduce.framework.name | local (default) | yarn | yarn |
| YARN | yarn.resourcemanager.hostname | N/A | localhost | resourcemanager |
| yarn.nodemanager.aux-services | N/A | mapreduce_shuffle | mapreduce_shuffle |
You can read more about configuration in Hadoop Configuration.
Standalone Mode
In standalone mode, there is no further action to take, since the default properties are set for standalone mode and there are no daemons to run.
Pseudodistributed Mode
In pseudodistributed mode, the configuration files should be created with the following contents and placed in the etc/hadoop directory. Alternatively, you can copy the etc/hadoop directory to another location, and then place the *-site.xml configuration files there. The advantage of this approach is that it separates configuration settings from the installation files. If you do this, you need to set the HADOOPCONF_DIR environment variable to the alternative location, or make sure you start the daemons with the —config option:
<?xml version=”1.0”?>
<?xml version=”1.0”?>
<?xml version=”1.0”?>
<?xml version=”1.0”?>
_
Configuring SSH
In pseudodistributed mode, we have to start daemons, and to do that using the supplied scripts we need to have SSH installed. Hadoop doesn’t actually distinguish between pseudodistributed and fully distributed modes; it merely starts daemons on the set of hosts in the cluster (defined by the slaves file) by SSHing to each host and starting a daemon process. Pseudodistributed mode is just a special case of fully distributed mode in which the (single) host is localhost, so we need to make sure that we can SSH to localhost and log in without having to enter a password.
First, make sure that SSH is installed and a server is running. On Ubuntu, for example, this is achieved with:
% sudo apt-get install ssh
Then, to enable passwordless login, generate a new SSH key with an empty passphrase:
% ssh-keygen -t rsa -P ‘’ -f ~/.ssh/id_rsa
% cat ~/.ssh/id_rsa.pub >> ~/.ssh/authorized_keys
You may also need to run ssh-add if you are running ssh-agent.
Test that you can connect with:
% ssh localhost
If successful, you should not have to type in a password.
Formatting the HDFS filesystem
Before HDFS can be used for the first time, the filesystem must be formatted. This is done by running the following command:
% hdfs namenode -format
Starting and stopping the daemons
To start the HDFS, YARN, and MapReduce daemons, type:
% start-dfs.sh
% start-yarn.sh
% mr-jobhistory-daemon.sh start historyserver
NOTE If you have placed configuration files outside the default conf directory, either export the HADOOPCONF_DIR environment variable before running the scripts, or start the daemons with the —config option, which takes an absolute path to the configuration directory:
If you have placed configuration files outside the default conf directory, either export the HADOOPCONF_DIR environment variable before running the scripts, or start the daemons with the —config option, which takes an absolute path to the configuration directory:
% **start-dfs.sh —config _path-to-config-directory
% start-yarn.sh —config path-to-config-directory
% mr-jobhistory-daemon.sh —config path-to-config-directory start historyserver
The following daemons will be started on your local machine: a namenode, a secondary namenode, a datanode (HDFS), a resource manager, a node manager (YARN), and a history server (MapReduce). You can check whether the daemons started successfully by looking at the logfiles in the logs directory (in the Hadoop installation directory) or by looking at the web UIs, at http://localhost:50070/ for the namenode, http://localhost:8088/ for the resource manager, and http://localhost:19888/ for the history server. You can also use Java’s jps command to see whether the processes are running.
Stopping the daemons is done as follows:
% mr-jobhistory-daemon.sh stop historyserver
% stop-yarn.sh
% stop-dfs.sh**
Creating a user directory
Create a home directory for yourself by running the following:
% hadoop fs -mkdir -p /user/$USER
Fully Distributed Mode
Setting up a cluster of machines brings many additional considerations, so this mode is covered in Chapter 10.
Appendix B. Cloudera’s Distribution Including Apache Hadoop
Cloudera’s Distribution Including Apache Hadoop (hereafter CDH) is an integrated Apache Hadoop–based stack containing all the components needed for production, tested and packaged to work together. Cloudera makes the distribution available in a number of different formats: Linux packages, virtual machine images, tarballs, and tools for running CDH in the cloud. CDH is free, released under the Apache 2.0 license, and available at http://www.cloudera.com/cdh.
As of CDH 5, the following components are included, many of which are covered elsewhere in this book:
Apache Avro
A cross-language data serialization library; includes rich data structures, a fast/compact binary format, and RPC
Apache Crunch
A high-level Java API for writing data processing pipelines that can run on MapReduce or Spark
Apache DataFu (incubating)
A library of useful statistical UDFs for doing large-scale analyses
Apache Flume
Highly reliable, configurable streaming data collection
Apache Hadoop
Highly scalable data storage (HDFS), resource management (YARN), and processing (MapReduce)
Apache HBase
Column-oriented real-time database for random read/write access
Apache Hive
SQL-like queries and tables for large datasets
Hue
Web UI to make it easy to work with Hadoop data
Cloudera Impala
Interactive, low-latency SQL queries on HDFS or HBase
Kite SDK
APIs, examples, and docs for building apps on top of Hadoop
Apache Mahout
Scalable machine-learning and data-mining algorithms
Apache Oozie
Workflow scheduler for interdependent Hadoop jobs
Apache Parquet (incubating)
An efficient columnar storage format for nested data
Apache Pig
Data flow language for exploring large datasets
Cloudera Search
Free-text, Google-style search of Hadoop data
Apache Sentry (incubating)
Granular, role-based access control for Hadoop users
Apache Spark
A cluster computing framework for large-scale in-memory data processing in Scala, Java, and Python
Apache Sqoop
Efficient transfer of data between structured data stores (like relational databases) and
Hadoop
Apache ZooKeeper
Highly available coordination service for distributed applications
Cloudera also provides Cloudera Manager for deploying and operating Hadoop clusters running CDH.
To download CDH and Cloudera Manager, visit http://www.cloudera.com/downloads.
Appendix C. Preparing the NCDC Weather Data
This appendix gives a runthrough of the steps taken to prepare the raw weather datafiles so they are in a form that is amenable to analysis using Hadoop. If you want to get a copy of the data to process using Hadoop, you can do so by following the instructions given at the website that accompanies this book. The rest of this appendix explains how the raw weather datafiles were processed.
The raw data is provided as a collection of tar files, compressed with bzip2. Each year’s worth of readings comes in a separate file. Here’s a partial directory listing of the files:
1901.tar.bz2
1902.tar.bz2
1903.tar.bz2… 2000.tar.bz2
Each tar file contains a file for each weather station’s readings for the year, compressed with gzip. (The fact that the files in the archive are compressed makes the bzip2 compression on the archive itself redundant.) For example:
% tar jxf 1901.tar.bz2
% ls 1901 | head
029070-99999-1901.gz
029500-99999-1901.gz
029600-99999-1901.gz
029720-99999-1901.gz
029810-99999-1901.gz
227070-99999-1901.gz
Because there are tens of thousands of weather stations, the whole dataset is made up of a large number of relatively small files. It’s generally easier and more efficient to process a smaller number of relatively large files in Hadoop (see Small files and
CombineFileInputFormat), so in this case, I concatenated the decompressed files for a whole year into a single file, named by the year. I did this using a MapReduce program, to take advantage of its parallel processing capabilities. Let’s take a closer look at the program.
The program has only a map function. No reduce function is needed because the map does all the file processing in parallel with no combine stage. The processing can be done with a Unix script, so the Streaming interface to MapReduce is appropriate in this case; see Example C-1.
Example C-1. Bash script to process raw NCDC datafiles and store them in HDFS
#!/usr/bin/env bash
# NLineInputFormat gives a single line: key is offset, value is S3 URI _read offset s3file
# Retrieve file from S3 to local disk echo “reporter:status:Retrieving $s3file” >&2 $HADOOP_HOME/bin/hadoop fs -get $s3file .
# Un-bzip and un-tar the local file target=basename $s3file .tar.bz2 mkdir -p $target echo “reporter:status:Un-tarring $s3file to $target” >&2 tar jxf basename $s3file -C $target
# Un-gzip each station file and concat into one file echo “reporter:status:Un-gzipping $target” >&2 for file in $target// do
gunzip -c $file >> $target.all echo “reporter:status:Processed $file” >&2 done
# Put gzipped version into HDFS
echo “reporter:status:Gzipping $target and putting in HDFS” >&2 gzip -c $target.all | $HADOOP_HOME/bin/hadoop fs -put - gz/$target.gz
The input is a small text file (_ncdc_files.txt) listing all the files to be processed (the files start out on S3, so they are referenced using S3 URIs that Hadoop understands). Here is a sample:
s3n://hadoopbook/ncdc/raw/isd-1901.tar.bz2 s3n://hadoopbook/ncdc/raw/isd-1902.tar.bz2… s3n://hadoopbook/ncdc/raw/isd-2000.tar.bz2
Because the input format is specified to be NLineInputFormat, each mapper receives one line of input, which contains the file it has to process. The processing is explained in the script, but briefly, it unpacks the bzip2 file and then concatenates each station file into a single file for the whole year. Finally, the file is gzipped and copied into HDFS. Note the use of hadoop fs -put - to consume from standard input.
Status messages are echoed to standard error with a reporter:status prefix so that they get interpreted as MapReduce status updates. This tells Hadoop that the script is making progress and is not hanging.
The script to run the Streaming job is as follows:
% hadoop jar $HADOOP_HOME/share/hadoop/tools/lib/hadoop-streaming-*.jar \ -D mapred.reduce.tasks=0 \
-D mapred.map.tasks.speculative.execution=false \
-D mapred.task.timeout=12000000 \
-input ncdc_files.txt \
-inputformat org.apache.hadoop.mapred.lib.NLineInputFormat \
-output output \
-mapper load_ncdc_map.sh \ -file load_ncdc_map.sh
I set the number of reduce tasks to zero, since this is a map-only job. I also turned off speculative execution so duplicate tasks wouldn’t write the same files (although the approach discussed in Task side-effect files would have worked, too). The task timeout was set to a high value so that Hadoop doesn’t kill tasks that are taking a long time (for example, when unarchiving files or copying to HDFS, when no progress is reported).
Finally, the files were archived on S3 by copying them from HDFS using distcp.
Appendix D. The Old and New Java MapReduce APIs
The Java MapReduce API used throughout this book is called the “new API,” and it replaces the older, functionally equivalent API. Although Hadoop ships with both the old and new MapReduce APIs, they are not compatible with each other. Should you wish to use the old API, you can, since the code for all the MapReduce examples in this book is available for the old API on the book’s website (in the oldapi package).
There are several notable differences between the two APIs: The new API is in the org.apache.hadoop.mapreduce package (and subpackages).
The new API is in the org.apache.hadoop.mapreduce package (and subpackages).
The old API can still be found in org.apache.hadoop.mapred.
The new API favors abstract classes over interfaces, since these are easier to evolve. This means that you can add a method (with a default implementation) to an abstract class without breaking old implementations of the class.168] For example, the Mapper and Reducer interfaces in the old API are abstract classes in the new API.  The new API makes extensive use of context objects that allow the user code to communicate with the MapReduce system. The new Context, for example, essentially unifies the role of the JobConf, the OutputCollector, and the Reporter from the old API.
The new API makes extensive use of context objects that allow the user code to communicate with the MapReduce system. The new Context, for example, essentially unifies the role of the JobConf, the OutputCollector, and the Reporter from the old API. In both APIs, key-value record pairs are pushed to the mapper and reducer, but in addition, the new API allows both mappers and reducers to control the execution flow by overriding the run() method. For example, records can be processed in batches, or the execution can be terminated before all the records have been processed. In the old API, this is possible for mappers by writing a MapRunnable, but no equivalent exists for reducers.
In both APIs, key-value record pairs are pushed to the mapper and reducer, but in addition, the new API allows both mappers and reducers to control the execution flow by overriding the run() method. For example, records can be processed in batches, or the execution can be terminated before all the records have been processed. In the old API, this is possible for mappers by writing a MapRunnable, but no equivalent exists for reducers. Job control is performed through the Job class in the new API, rather than the old
Job control is performed through the Job class in the new API, rather than the old
JobClient, which no longer exists in the new API.
Configuration has been unified in the new API. The old API has a special JobConf object for job configuration, which is an extension of Hadoop’s vanilla Configuration object (used for configuring daemons; see The Configuration API). In the new API, job configuration is done through a Configuration, possibly via some of the helper methods on Job. Output files are named slightly differently: in the old API both map and reduce outputs are named part-__nnnnn, whereas in the new API map outputs are named part-m-nnnnn _and reduce outputs are named _part-r-nnnnn (where nnnnn is an integer designating the part number, starting from 00000).
Output files are named slightly differently: in the old API both map and reduce outputs are named part-__nnnnn, whereas in the new API map outputs are named part-m-nnnnn _and reduce outputs are named _part-r-nnnnn (where nnnnn is an integer designating the part number, starting from 00000). User-overridable methods in the new API are declared to throw java.lang.InterruptedException. This means that you can write your code to be responsive to interrupts so that the framework can gracefully cancel long-running operations if it needs to.169]
User-overridable methods in the new API are declared to throw java.lang.InterruptedException. This means that you can write your code to be responsive to interrupts so that the framework can gracefully cancel long-running operations if it needs to.169] In the new API, the reduce() method passes values as a java.lang.Iterable, rather than a java.lang.Iterator (as the old API does). This change makes it easier to iterate over the values using Java’s for-each loop construct:
In the new API, the reduce() method passes values as a java.lang.Iterable, rather than a java.lang.Iterator (as the old API does). This change makes it easier to iterate over the values using Java’s for-each loop construct:
for (VALUEIN value : values) { … }
WARNING Programs using the new API that were compiled against Hadoop 1 need to be recompiled to run against Hadoop 2. This is because some classes in the new MapReduce API changed to interfaces between the Hadoop 1 and Hadoop 2 releases. The symptom is an error at runtime like the following:
Programs using the new API that were compiled against Hadoop 1 need to be recompiled to run against Hadoop 2. This is because some classes in the new MapReduce API changed to interfaces between the Hadoop 1 and Hadoop 2 releases. The symptom is an error at runtime like the following:
java.lang.IncompatibleClassChangeError: Found interface org.apache.hadoop.mapreduce.TaskAttemptContext, but class was expected
Example D-1 shows the MaxTemperature application (from Java MapReduce) rewritten to use the old API. The differences are highlighted in bold.
WARNING When converting your Mapper and Reducer classes to the new API, don’t forget to change the signatures of the map() and reduce() methods to the new form. Just changing your class to extend the new Mapper or Reducer classes will not produce a compilation error or warning, because these classes provide identity forms of the map() and reduce() methods (respectively). Your mapper or reducer code, however, will not be invoked, which can lead to some hard-todiagnose errors.
When converting your Mapper and Reducer classes to the new API, don’t forget to change the signatures of the map() and reduce() methods to the new form. Just changing your class to extend the new Mapper or Reducer classes will not produce a compilation error or warning, because these classes provide identity forms of the map() and reduce() methods (respectively). Your mapper or reducer code, however, will not be invoked, which can lead to some hard-todiagnose errors.
Annotating your map() and reduce() methods with the @Override annotation will allow the Java compiler to catch these errors.
Example D-1. Application to find the maximum temperature, using the old MapReduce API
public class OldMaxTemperature {
static class OldMaxTemperatureMapper extends MapReduceBase implements Mapper
private static final int MISSING = 9999;
@Override
public void map(LongWritable key, Text value,
OutputCollector
String line = value.toString(); String year = line.substring(15, 19); int airTemperature; if (line.charAt(87) == ‘+’) { // parseInt doesn’t like leading plus signs airTemperature = Integer.parseInt(line.substring(88, 92));
} else {
airTemperature = Integer.parseInt(line.substring(87, 92));
}
String quality = line.substring(92, 93);
if (airTemperature != MISSING && quality.matches(“[01459]”)) { output.collect(new Text(year), new IntWritable(airTemperature));
}
}
}
static class OldMaxTemperatureReducer extends MapReduceBase implements Reducer
@Override
public void reduce(Text key, Iterator
int maxValue = Integer.MIN_VALUE; while (values.hasNext()) { maxValue = Math.max(maxValue, values.next().get());
}
output.collect(key, new IntWritable(maxValue));
} } public static void main(String[] args) throws IOException { if (args.length != 2) {
System.err.println(“Usage: OldMaxTemperature
Index
A
AbstractAvroEventSerializer class, File Formats access control lists (ACLs), An example, ACLs accumulators, Accumulators
ACLs (access control lists), An example, ACLs action nodes, Apache Oozie, Defining an Oozie workflow actions, RDD, Transformations and Actions
ADAM platform, ADAM, A Scalable Genome Analysis Platform–A simple example: kmer counting using Spark and ADAM ADMIN permission (ACL), ACLs
administration (see system administration) agents (Flume) about, Flume
distribution process, Distribution: Agent Tiers–Delivery Guarantees example of, An Example–An Example HDFS sinks and, The HDFS Sink Aggregate class, Crunch Libraries
aggregating data in Hive tables, Sorting and Aggregating
Aggregator interface, An Example
Aggregators class, An Example, combineValues() aliases, defining, A Filter UDF
ALTER TABLE statement (Hive), Partitions, Altering Tables
Amazon Web Services, Data!, A Brief History of Apache Hadoop
Ant build tool, Setting Up the Development Environment, Packaging a Job
Apache Avro (see Avro)
Apache Commons Logging API, Hadoop Logs
Apache Crunch (see Crunch)
Apache Curator project, More Distributed Data Structures and Protocols
Apache Flume (see Flume)
Apache Mesos, Executors and Cluster Managers
Apache Oozie
about, Apache Oozie
defining workflows, Defining an Oozie workflow–Defining an Oozie workflow packaging and deploying workflow applications, Packaging and deploying an Oozie workflow application
running workflow job, Running an Oozie workflow job
Apache Parquet (see Parquet)
Apache Phoenix, SQL-on-Hadoop Alternatives
Apache Slider, Application Lifespan
Apache Software Foundation, Beyond Batch, Building YARN Applications
Apache Spark (see Spark)
Apache Tez, Execution engines
Apache Thrift, Serialization IDL
Apache Twill, Building YARN Applications APPEND write mode, Existing outputs application IDs, Launching a Job application masters about, Anatomy of a YARN Application Run decommissioning old nodes, Decommissioning old nodes failure considerations, Application Master Failure job completion, Job Completion job initialization process, Job Initialization jobtrackers and, YARN Compared to MapReduce 1 node manager failure, Node Manager Failure progress and status updates, Progress and Status Updates resource manager failure, Resource Manager Failure task assignments, Task Assignment task execution, Task Execution task failure, Task Failure
unmanaged, Anatomy of a YARN Application Run
ArrayFile class, MapFile variants
ArrayPrimitiveWritable class, Writable collections
ArrayWritable class, Writable collections
ASSERT statement (Pig Latin), Statements
Astrometry.net project, Data!
atomic broadcast, Implementation
audit logs (HDFS), Hadoop Logs, Audit Logging authentication
ACLs and, ACLs
delegation tokens and, Delegation Tokens
Kerberos, Kerberos and Hadoop–An example, ACLs authorization process, Kerberos and Hadoop
AVG function (Pig Latin), Functions
Avro about, Serialization IDL, Avro–Avro
binary storage format and, Binary storage formats: Sequence files, Avro datafiles, Parquet files, RCFiles, and ORCFiles Crunch and, Types
data types and schemas, Avro Data Types and Schemas–Avro Data Types and Schemas
datafile format, Other File Formats and Column-Oriented Formats, Avro Datafiles–Avro Datafiles
Flume support, File Formats
Hive support, Text and Binary File Formats interoperability, Interoperability–Avro Tools
languages and framework support, Avro in Other Languages
MapReduce support, Avro MapReduce–Sorting Using Avro MapReduce Parquet and, Avro, Protocol Buffers, and Thrift–Projection and read schemas schema resolution, Schema Resolution–Schema Resolution
serialization and deserialization, In-Memory Serialization and Deserialization–The Specific API
sort capabilities, Sort Order, Sorting Using Avro MapReduce–Sorting Using Avro MapReduce
Sqoop support, Text and Binary File Formats tools supported, Avro Tools
avro.java.string property, Avro Data Types and Schemas
AvroAsTextInputFormat class, Avro in Other Languages
AvroParquetOutputFormat class, Parquet MapReduce
AvroParquetReader class, Avro, Protocol Buffers, and Thrift
AvroParquetWriter class, Avro, Protocol Buffers, and Thrift
AvroReadSupport class, Projection and read schemas AvroStorage function (Pig Latin), Functions
awk tool, Analyzing the Data with Unix Tools–Analyzing the Data with Unix Tools, Running a Distributed MapReduce Job
B
B-Tree data structure, Relational Database Management Systems backups data, Data backups metadata, Metadata backups
balancer tool, Keeping an HDFS Cluster Balanced, Balancer, Filesystem balancer Baldeschwieler, Eric, A Brief History of Apache Hadoop bandwidth, measuring between nodes, Anatomy of a File Read batch processing
Flume support, Batching limitations of, Beyond Batch
batchSize property, Batching Beeline service (Hive), Hive Services
benchmarking clusters, Benchmarking a Hadoop Cluster–User Jobs binary formats for data storage, Binary storage formats: Sequence files, Avro datafiles, Parquet files, RCFiles, and ORCFiles
FixedLengthInputFormat class, FixedLengthInputFormat
MapFileOutputFormat class, MapFileOutputFormat
SequenceFileAsBinaryInputFormat class, SequenceFileAsBinaryInputFormat
SequenceFileAsBinaryOutputFormat class, SequenceFileAsBinaryOutputFormat
SequenceFileAsTextInputFormat class, SequenceFileAsTextInputFormat
SequenceFileInputFormat class, SequenceFileInputFormat
SequenceFileOutputFormat class, SequenceFileOutputFormat biological data science case study
about, Biological Data Science: Saving Lives with Software–Biological Data Science: Saving Lives with Software
ADAM platform, ADAM, A Scalable Genome Analysis Platform–A simple example: kmer counting using Spark and ADAM
DNA as source code, Thinking of DNA as Source Code–Thinking of DNA as Source
Code
genetic code, The Genetic Code: Turning DNA Letters into Proteins
Human Genome Project, The Human Genome Project and Reference Genomes join in, Join In
personalized medicine and, From Personalized Ads to Personalized Medicine–From Personalized Ads to Personalized Medicine
reference genomes, The Human Genome Project and Reference Genomes sequencing and aligning DNA, Sequencing and Aligning DNA structure of DNA, The Structure of DNA
blacklisting node managers, Node Manager Failure block access tokens, Delegation Tokens
blockpoolID identifier, Namenode directory structure blocks and block sizes about, Blocks–Blocks block caching, Block Caching
checking blocks, Filesystem check (fsck)–Finding the blocks for a file input splits and, TextInputFormat Parquet and, Parquet Configuration setting, HDFS block size
setting for HDFS, FileInputFormat input splits
BloomMapFile class, MapFile variants
BookKeeper service, BookKeeper and Hedwig
BooleanWritable class, Writable wrappers for Java primitives broadcast variables, Broadcast Variables Brush, Ryan, Composable Data at Cerner
buckets, Hive tables and, Partitions and Buckets, Buckets–Buckets buffer size, Buffer size built-in counters
about, Built-in Counters job counters, Job counters
task counters, Built-in Counters–Task counters
bulk loading, Bulk load
ByteArrayOutputStream class (Java), The Writable Interface
BytesWritable class, BytesWritable, Processing a whole file as a record
BYTES_READ counter, Task counters
BYTES_WRITTEN counter, Task counters
ByteWritable class, Writable wrappers for Java primitives bzip2 compression, Compression–Codecs, Native libraries
BZip2Codec class, Codecs
C
C library (libhdfs), C
Cafarella, Mike, A Brief History of Apache Hadoop, Backdrop
Capacity Scheduler (YARN), Capacity Scheduler Configuration–Queue placement
Cartesian class, Crunch Libraries
Cascading library case study about, Cascading
application example, Cascading in Practice–Cascading in Practice fields, tuples, and pipes, Fields, Tuples, and Pipes–Fields, Tuples, and Pipes flexibility, Flexibility operations, Operations–Operations
ShareThis sharing network, Hadoop and Cascading at ShareThis–Hadoop and
Cascading at ShareThis summary, Summary
taps, schemes, and flows, Taps, Schemes, and Flows
case sensitivity
HiveQL, The Hive Shell, Writing a UDF
Pig Latin, Structure case studies
biological data science, Biological Data Science: Saving Lives with Software–Join In Cascading library, Cascading–Summary
composable data at Cerner, Composable Data at Cerner–Moving Forward
cat command, Statements cd command, Statements
CDH (Cloudera’s Distribution including Apache Hadoop), Cloudera’s Distribution Including Apache Hadoop–Cloudera’s Distribution Including Apache Hadoop
Cerner case study, Composable Data at Cerner about, From CPUs to Semantic Integration Apache Crunch and, Enter Apache Crunch
building complete picture, Building a Complete Picture–Integrating Healthcare Data composability over frameworks, Composability over Frameworks
integrating healthcare data, Integrating Healthcare Data–Integrating Healthcare Data moving forward, Moving Forward
ChainMapper class, Decomposing a Problem into MapReduce Jobs, MapReduce Library Classes
ChainReducer class, Decomposing a Problem into MapReduce Jobs, MapReduce Library Classes
channel property, An Example Channels class, Crunch Libraries channels property, An Example
CharSequence interface (Java), Avro Data Types and Schemas
CHECKPOINT write mode, Existing outputs, Checkpointing a Pipeline checkpointing process running against namenodes, The filesystem image and edit log running against pipelines, Checkpointing a Pipeline
ChecksumFileSystem class, Data Integrity, ChecksumFileSystem checksumming data, Data Integrity–ChecksumFileSystem clear command, Statements
Closeable interface (Java), Writing a SequenceFile
CLOSED state (ZooKeeper), States
Cloudera’s Distribution including Apache Hadoop (CDH), Cloudera’s Distribution
Including Apache Hadoop–Cloudera’s Distribution Including Apache Hadoop
CLUSTER BY clause (Hive), Sorting and Aggregating cluster managers, Executors and Cluster Managers CLUSTERED BY clause (Hive), Buckets clusterID identifier, Namenode directory structure ClusterMapReduceTestCase class, Testing the Driver clusters administration tools and, dfsadmin–Balancer audit logging and, Audit Logging
balancing, Keeping an HDFS Cluster Balanced benchmarking, Benchmarking a Hadoop Cluster–User Jobs
Hadoop configuration additional properties, Other Hadoop Properties configuration files, Hadoop Configuration configuration management, Configuration Management
daemon addresses and ports, Hadoop Daemon Addresses and Ports–Hadoop Daemon Addresses and Ports
daemon properties, Important Hadoop Daemon Properties–CPU settings in YARN and MapReduce
environment variables, Hadoop Configuration, Environment Settings–SSH settings
maintenance considerations, Metadata backups–Finalize the upgrade (optional) monitoring, Monitoring–Metrics and JMX network topology, Network Topology–Rack awareness
persistent data structures and, Persistent Data Structures–Datanode directory structure running MapReduce applications on, Running on a Cluster–Remote Debugging safe mode and, Safe Mode–Entering and leaving safe mode security considerations, Security–Other Security Enhancements setup and installation
configuring Hadoop, Configuring Hadoop configuring SSH, Configuring SSH
creating Unix user accounts, Creating Unix User Accounts creating user directories, Creating User Directories formatting HDFS filesystem, Formatting the HDFS Filesystem installation options, Setting Up a Hadoop Cluster installing Hadoop, Installing Hadoop installing Java, Installing Java
starting and stopping daemons, Starting and Stopping the Daemons–Starting and
Stopping the Daemons sizing, Cluster Sizing–Master node scenarios specifications for, Cluster Specification–Rack awareness testing in miniclusters, Testing the Driver upgrading, Upgrades–Finalize the upgrade (optional) CodecPool class, CodecPool codecs about, Codecs
compressing streams, Compressing and decompressing streams with CompressionCodec
decompressing streams, Compressing and decompressing streams with CompressionCodec
inferring from file extensions, Inferring CompressionCodecs using CompressionCodecFactory
list of supported compression formats, Codecs native libraries, Native libraries–CodecPool
codegen tool, Generated Code, Exports and SequenceFiles
Cogroup class, Crunch Libraries
COGROUP statement (Pig Latin), Statements, COGROUP–COGROUP
coherency models (filesystems), Coherency Model–Consequences for application design Collection interface (Java), PObject column-oriented storage about, Other File Formats and Column-Oriented Formats–Other File Formats and Column-Oriented Formats
Parquet and, Parquet–Parquet MapReduce
com.sun.management.jmxremote.port property, Metrics and JMX CombineFileInputFormat class, Small files and CombineFileInputFormat combiner functions about, Combiner Functions–Specifying a combiner function general form, MapReduce Types shuffle and sort, The Reduce Side tuning checklist, Tuning a Job
COMBINE_INPUT_RECORDS counter, Task counters COMBINE_OUTPUT_RECORDS counter, Task counters command-line interface about, The Command-Line Interface–Basic Filesystem Operations
displaying SequenceFile with, Displaying a SequenceFile with the command-line interface
Hive support, Hive Services–Hive clients Pig Latin support, Statements
running MapReduce jobs from, GenericOptionsParser, Tool, and ToolRunner–GenericOptionsParser, Tool, and ToolRunner running miniclusters from, Testing the Driver
ZooKeeper support, ZooKeeper command-line tools comments (Pig Latin), Structure
commissioning nodes, Commissioning and Decommissioning Nodes–Commissioning new nodes
COMMITTED_HEAP_BYTES counter, Task counters, Memory settings in YARN and MapReduce
Comparable interface (Java), WritableComparable and comparators Comparator interface (Java), WritableComparable and comparators compatibility, upgrades and, Upgrades CompositeInputFormat class, Map-Side Joins compression
Compression codecs and, Codecs–CodecPool
input splits and, Compression and Input Splits list of supported formats, Compression map output and, The Map Side
MapReduce and, Compression and Input Splits–Compressing map output Parquet and, Parquet File Format
selecting format to use, Compression and Input Splits SequenceFiles, The SequenceFile format tuning checklist, Tuning a Job
CompressionCodec interface about, Codecs
compressing streams, Compressing and decompressing streams with CompressionCodec
decompressing streams, Compressing and decompressing streams with CompressionCodec
inferring codecs, Inferring CompressionCodecs using
CompressionCodecFactory–Inferring CompressionCodecs using CompressionCodecFactory
CompressionCodecFactory class, Inferring CompressionCodecs using CompressionCodecFactory
CompressionInputStream class, Compressing and decompressing streams with CompressionCodec
CompressionOutputStream class, Compressing and decompressing streams with
CompressionCodec
CONCAT function (Pig Latin), Functions
conf command (ZooKeeper), Installing and Running ZooKeeper
Configurable interface, GenericOptionsParser, Tool, and ToolRunner
Configuration class about, Reading Data Using the FileSystem API, The Configuration API–The
Configuration API, GenericOptionsParser, Tool, and ToolRunner combining resources, Combining Resources side data distribution, Using the Job Configuration variable expansion, Variable Expansion configuration files, listed, Hadoop Configuration
Configured class, GenericOptionsParser, Tool, and ToolRunner
CONNECTED state (ZooKeeper), States
CONNECTING state (ZooKeeper), States
Connection interface, Java
ConnectionDriverName class, The Metastore
ConnectionFactory class, Java
ConnectionPassword class, The Metastore
ConnectionURL class, The Metastore ConnectionUserName class, The Metastore connectors (Sqoop), Sqoop Connectors ConnectTimeout SSH setting, SSH settings
cons command (ZooKeeper), Installing and Running ZooKeeper containers about, Anatomy of a YARN Application Run jobtrackers and, YARN Compared to MapReduce 1
virtual memory constraints, Memory settings in YARN and MapReduce
control flow statements, Statements
control-flow nodes, Apache Oozie, Defining an Oozie workflow converting Hive data types, Conversions coordinator engines, Apache Oozie copyFromLocal command, Statements copyToLocal command, Statements
core-site.xml file, Hadoop Configuration, Important Hadoop Daemon Properties, An example
COUNT function (Pig Latin), Functions counters
Counters built-in, Built-in Counters–Task counters Crunch support, Running a Pipeline dynamic, Dynamic counters HBase and, Counters metrics and, Metrics and JMX
retrieving, Retrieving counters–Retrieving counters
user-defined Java, User-Defined Java Counters–Retrieving counters user-defined Streaming, User-Defined Streaming Counters
Counters class, Retrieving counters, MapReduce COUNT_STAR function (Pig Latin), Functions cp command, Statements
CPU_MILLISECONDS counter, Task counters
CRC-32 (cyclic redundancy check), Data Integrity
CREATE DATABASE statement (Hive), Tables
CREATE FUNCTION statement (Hive), Writing a UDF create operation (ZooKeeper), Operations
CREATE permission (ACL), ACLs
CREATE TABLE statement (Hive), An Example, Managed Tables and External Tables, Binary storage formats: Sequence files, Avro datafiles, Parquet files, RCFiles, and ORCFiles
CREATE TABLE…AS SELECT statement (Hive), CREATE TABLE…AS SELECT
Crick, Francis, The Structure of DNA
CROSS statement (Pig Latin), Statements, CROSS crst command (ZooKeeper), Installing and Running ZooKeeper
Crunch
Crunch
Cerner case study and, Enter Apache Crunch functions, Functions–Object reuse
libraries supported, Crunch Libraries–Crunch Libraries materialization process, Materialization–PObject
pipeline execution, Pipeline Execution–Inspecting a Crunch Plan primitive operations, Primitive Operations–combineValues() records and tuples, Records and tuples–Records and tuples sorting data, Total Sort
sources and targets, Sources and Targets–Combined sources and targets types supported, Types–Records and tuples weather dataset example, An Example–An Example
crunch.log.job.progress property, Debugging cTime property, Namenode directory structure
CUBE statement (Pig Latin), Statements
Cutting, Doug, A Brief History of Apache Hadoop–A Brief History of Apache Hadoop,
Avro
cyclic redundancy check (CRC-32), Data Integrity
D
daemons addresses and ports, Hadoop Daemon Addresses and Ports–Hadoop Daemon Addresses and Ports
balancer tool, Balancer
configuration properties, Important Hadoop Daemon Properties–CPU settings in YARN and MapReduce
logfile support, Hadoop Logs, System logfiles, Logging memory requirements, Memory heap size
starting and stopping, Starting and Stopping the Daemons–Starting and Stopping the Daemons, Starting and stopping the daemons
DAGs (directed acyclic graphs), JobControl, Pipeline Execution, DAG
Construction–DAG Construction
data integrity
Data Integrity
ChecksumFileSystem class, ChecksumFileSystem
HDFs support, Data Integrity in HDFS
LocalFileSystem class, LocalFileSystem data local tasks, Task Assignment data locality, Grid Computing data locality optimization, Data Flow data queue, Anatomy of a File Write data storage and analysis about, Data Storage and Analysis
analyzing data with Hadoop, Analyzing the Data with Hadoop–A test run
analyzing data with Unix tools, Analyzing the Data with Unix Tools–Analyzing the Data with Unix Tools
column-oriented formats, Other File Formats and Column-Oriented Formats–Other File Formats and Column-Oriented Formats
HDFS blocks and, Blocks
Hive tables, Storage Formats–Storage handlers
scaling out, Scaling Out–Running a Distributed MapReduce Job system comparisons, Comparison with Other Systems–Volunteer Computing
data structures additional formats, Other File Formats and Column-Oriented Formats–Other File Formats and Column-Oriented Formats MapFile class, MapFile
persistent, Persistent Data Structures–Datanode directory structure SequenceFile class, SequenceFile–The SequenceFile format
ZooKeeper and, More Distributed Data Structures and Protocols database formats, Database Input (and Output) DataBlockScanner class, Data Integrity in HDFS dataDir property, Resilience and Performance
DataDrivenDBInputFormat class, Imports: A Deeper Look
DataFileReader class, Avro Datafiles
DataFileStream class, Avro Datafiles
DataFileWriter class, Avro Datafiles
DataInput interface (Java), The Writable Interface dataLogDir property, Resilience and Performance DataNodeProtocol interface, Other Security Enhancements datanodes balancer tool and, Balancer
block scanners and, Datanode block scanner
cluster setup and installation, Formatting the HDFS Filesystem
commissioning nodes, Commissioning new nodes–Commissioning new nodes data integrity and, Data Integrity in HDFS DataStreamer class and, Anatomy of a File Write
decommissioning nodes, Decommissioning old nodes–Decommissioning old nodes DFSInputStream class and, Anatomy of a File Read directory structure, Datanode directory structure HBase and, HDFS
master−worker pattern, Namenodes and Datanodes RAID storage and, Cluster Specification replica placement, Anatomy of a File Write starting, Starting and Stopping the Daemons
DataOutput interface (Java), The Writable Interface
DataOutputStream class (Java), The Writable Interface
DataStreamer class, Anatomy of a File Write–Anatomy of a File Write
DATA_LOCAL_MAPS counter, Job counters
DatumWriter interface, In-Memory Serialization and Deserialization
DBInputFormat class, Input Splits and Records
DBOutputFormat class, Database Input (and Output) DBWritable interface, Imports: A Deeper Look debugging problems
Debugging a Job–Debugging a Job Crunch and, Debugging
handling malformed data, Handling malformed data
MapReduce task attempts page, The tasks and task attempts pages MapReduce tasks page, The tasks and task attempts pages remotely, Remote Debugging
setting log levels, Setting log levels
decommissioning nodes, Decommissioning old nodes–Decommissioning old nodes
DefaultCodec class, Codecs
DefaultStringifier class, Using the Job Configuration
DEFINE statement (Pig Latin), Statements, A Filter UDF, STREAM
DEFLATE compression, Compression–Codecs, Native libraries
DeflaterInputStream class (Java), Compressing and decompressing streams with CompressionCodec
DeflaterOutputStream class (Java), Compressing and decompressing streams with
CompressionCodec
delay scheduling, Delay Scheduling delegation tokens, Delegation Tokens delete operation (ZooKeeper), Operations
DELETE permission (ACL), ACLs
DELETE statement (Hive), Updates, Transactions, and Indexes
DELIMITED keyword (Hive), Using a custom SerDe: RegexSerDe
delimited text storage format, The default storage format: Delimited text–The default storage format: Delimited text
dependencies, job, Packaging dependencies
DESCRIBE operator (Pig Latin), An Example, Comparison with Databases, Statements, Anonymous Relations
DESCRIBE statement (Hive), Operators and Functions, Views deserialization about, Serialization
Avro support, In-Memory Serialization and Deserialization–The Specific API column-oriented storage and, Other File Formats and Column-Oriented Formats
Text class and, Implementing a Custom Writable
Deserializer interface, Serialization Frameworks development environment managing configuration, Managing Configuration–Managing Configuration running jobs from command-line, GenericOptionsParser, Tool, and ToolRunner–GenericOptionsParser, Tool, and ToolRunner
setting up, Setting Up the Development Environment–Setting Up the Development
Environment df tool, Blocks
dfs.block.access.token.enable property, Delegation Tokens dfs.blocksize property, FileInputFormat input splits, HDFS block size dfs.bytes-per-checksum property, Data Integrity in HDFS dfs.client.read.shortcircuit property, Short-circuit local reads dfs.datanode.address property, Hadoop Daemon Addresses and Ports dfs.datanode.balance.bandwidthPerSec property, Balancer dfs.datanode.data.dir property, HDFS, YARN
dfs.datanode.http.address property, Hadoop Daemon Addresses and Ports dfs.datanode.ipc.address property, Hadoop Daemon Addresses and Ports dfs.datanode.kerberos.principal property, Other Security Enhancements dfs.datanode.keytab.file property, Other Security Enhancements dfs.datanode.numblocks property, Datanode directory structure dfs.datanode.scan.period.hours property, Datanode block scanner dfs.domain.socket.path property, Short-circuit local reads dfs.encrypt.data.transfer property, Other Security Enhancements dfs.hosts property, Cluster membership, Commissioning new nodes
dfs.namenode.checkpoint.dir property, HDFS, Secondary namenode directory structure dfs.namenode.checkpoint.period property, The filesystem image and edit log dfs.namenode.checkpoint.txns property, The filesystem image and edit log dfs.namenode.http-address property, Hadoop Daemon Addresses and Ports dfs.namenode.http-bind-host property, Hadoop Daemon Addresses and Ports dfs.namenode.name.dir property, HDFS, Namenode directory structure, Secondary namenode directory structure
dfs.namenode.replication.min property, Safe Mode
dfs.namenode.rpc-bind-host property, Hadoop Daemon Addresses and Ports
dfs.namenode.safemode.extension property, Safe Mode dfs.namenode.safemode.threshold-pct property, Safe Mode
dfs.namenode.secondary.http-address property, Hadoop Daemon Addresses and Ports dfs.permissions.enabled property, Basic Filesystem Operations dfs.replication property, The Command-Line Interface, Configuration dfs.webhdfs.enabled property, HTTP
dfsadmin tool, Entering and leaving safe mode–dfsadmin, Metadata backups
DFSInputStream class, Anatomy of a File Read
DFSOutputStream class, Anatomy of a File Write–Anatomy of a File Write DIFF function (Pig Latin), Functions digital universe, Data!–Data!
direct-mode imports, Direct-Mode Imports
directed acyclic graphs (DAGs), JobControl, Pipeline Execution, DAG
Construction–DAG Construction directories creating, Directories, Creating User Directories, Creating a user directory datanode structure, Datanode directory structure file permissions, Basic Filesystem Operations namenode memory requirements, The Design of HDFS
namenode structure, Namenode directory structure–Namenode directory structure querying, File metadata: FileStatus–PathFilter reserved storage space, Reserved storage space
secondary namenode structure, Secondary namenode directory structure
distcp program, Parallel Copying with distcp–Keeping an HDFS Cluster Balanced, Data Integrity in HDFS
Distinct class, Crunch Libraries
DISTINCT statement (Pig Latin), Statements, FOREACH…GENERATE DISTRIBUTE BY clause (Hive), Sorting and Aggregating distributed cache, Distributed Cache–The distributed cache API
DistributedCache class, The distributed cache API
DistributedFileSystem class
about, Hadoop Filesystems
FileSystem class and, The Java Interface HTTP support, HTTP
reading files, Anatomy of a File Read writing files, Anatomy of a File Write
DNA
ADAM platform, ADAM, A Scalable Genome Analysis Platform–A simple example: kmer counting using Spark and ADAM
genetic code, The Genetic Code: Turning DNA Letters into Proteins
Human Genome Project, The Human Genome Project and Reference Genomes sequencing and aligning, Sequencing and Aligning DNA
as source code, Thinking of DNA as Source Code–Thinking of DNA as Source Code structure of, The Structure of DNA
DNSToSwitchMapping interface, Rack awareness
DoFn class about, An Example
increment() method, Running a Pipeline scaleFactor() method, parallelDo()
Dominant Resource Fairness (DRF), Dominant Resource Fairness dot tool, Inspecting a Crunch Plan
DoubleWritable class, Writable wrappers for Java primitives
Dreadnaught, A Brief History of Apache Hadoop
DRF (Dominant Resource Fairness), Dominant Resource Fairness
DROP DATABASE statement (Hive), Tables
DROP FUNCTION statement (Hive), Writing a UDF
DROP TABLE statement (Hive), Managed Tables and External Tables, Dropping Tables dsh shell tool, Configuration Management Dumbo module (Python), Python
dump command (ZooKeeper), Installing and Running ZooKeeper
DUMP statement (Pig Latin), Statements, Statements, Sorting Data, Anonymous Relations Dyer, Chris, MapReduce Workflows dynamic counters, Dynamic counters dynamic-partition insert, Inserts
E
EC2 computer cloud, A Brief History of Apache Hadoop
edit log, The filesystem image and edit log–The filesystem image and edit log edits files, Namenode directory structure–The filesystem image and edit log, Metadata backups
Elephant Bird project, Serialization IDL embedded metastore, The Metastore
EmbeddedAgent class, Integrating Flume with Applications Encoder class, In-Memory Serialization and Deserialization encoding, nested, Nested Encoding
EnumSetWritable class, Writable collections
envi command (ZooKeeper), Installing and Running ZooKeeper
environment variables, A test run, Hadoop Configuration, Environment Settings–SSH settings
ephemeral znodes, Ephemeral znodes escape sequences, Performing an Export
eval functions (Pig Latin), Functions, An Eval UDF–Dynamic invokers EvalFunc class, A Filter UDF
exceptions (ZooKeeper), The Resilient ZooKeeper Application–A reliable configuration service, Recoverable exceptions exec command, Statements, Statements
execute (x) permission, Basic Filesystem Operations exists operation (ZooKeeper), Operations EXPLAIN keyword (Hive), Inner joins EXPLAIN operator (Pig Latin), Statements export process (Sqoop) about, Performing an Export–Exports: A Deeper Look
Hive and, Performing an Export
SequenceFile and, Exports and SequenceFiles transactionality and, Exports and Transactionality
expressions (Pig Latin), Expressions–Expressions
EXTERNAL keyword (Hive), Managed Tables and External Tables
external tables (Hive), Managed Tables and External Tables–Managed Tables and External Tables
F
FAILED_SHUFFLE counter, Task counters
failover controllers, Failover and fencing, Resource Manager Failure failovers sink groups and, Sink Groups–Integrating Flume with Applications
ZooKeeper service and, Sessions failures about, Failures
application master, Application Master Failure node manager, Node Manager Failure resource manager, Resource Manager Failure task, Task Failure
Fair Scheduler (YARN), Fair Scheduler Configuration–Preemption fanning out about, Fan Out
delivery guarantees, Delivery Guarantees
replicating and multiplexing selectors, Replicating and Multiplexing Selectors
FieldSelectionMapper class, MapReduce Library Classes
FieldSelectionMapReduce class, MapReduce Library Classes
FieldSelectionReducer class, MapReduce Library Classes
FIFO Scheduler (YARN), Scheduler Options–Scheduler Options File class (Java), Directories file management
compression, Compression–Compressing map output, The SequenceFile format file patterns, File patterns
file permissions, Basic Filesystem Operations
file-based data structures, File-Based Data Structures–Other File Formats and ColumnOriented Formats
finding file checksum, Data Integrity in HDFS listing files, Listing files
Parquet considerations, Writing and Reading Parquet Files–Projection and read schemas processing files as records, Processing a whole file as a record–Processing a whole file as a record
file.bytes-per-checksum property, LocalFileSystem
FileContext class, The Java Interface
FileFilter class (Java), PathFilter
FileInputFormat class, FileInputFormat–FileInputFormat input splits, Built-in Counters, Reading from a source
FileOutputCommitter class, Job Initialization, Output Committers
FileOutputFormat class, Using Compression in MapReduce, Task side-effect files, Built-in Counters
FileSplit class, Processing a whole file as a record
FileStatus class, File metadata: FileStatus–File patterns
FileSystem class about, Hadoop Filesystems, Interfaces, The Java Interface creating directories, Directories deleting data, Deleting Data
querying directories, File metadata: FileStatus–PathFilter
reading data, Reading Data Using the FileSystem API–FSDataInputStream, Anatomy of a File Read
verifying checksums, Data Integrity in HDFS writing data, Writing Data–FSDataOutputStream Filesystem in Userspace (FUSE), FUSE filesystems
basic concepts, Blocks–Failover and fencing
basic operations, Basic Filesystem Operations–Basic Filesystem Operations built-in counters, Built-in Counters, Task counters
checking blocks, Filesystem check (fsck)–Finding the blocks for a file cluster sizing, Master node scenarios
coherency models, Coherency Model–Consequences for application design formatting for HDFS, Formatting the HDFS Filesystem Hadoop supported, Hadoop Filesystems–FUSE
high availability, HDFS High Availability–Failover and fencing Java interface, The Java Interface–Deleting Data
metadata and, The filesystem image and edit log–The filesystem image and edit log namenodes and, The Design of HDFS, Namenodes and Datanodes
parallel copying with distcp, Parallel Copying with distcp–Keeping an HDFS Cluster Balanced
upgrade considerations, Upgrades–Finalize the upgrade (optional) FileUtil class, Listing files
filter functions (Pig Latin), Functions, A Filter UDF–Leveraging types
FILTER statement (Pig Latin), Statements, FOREACH…GENERATE
FilterFn class, parallelDo()
FilterFunc class, A Filter UDF
FirstKeyOnlyFilter class, MapReduce
FixedLengthInputFormat class, FixedLengthInputFormat
FloatWritable class, Writable wrappers for Java primitives
Flume
about, Flume, Crunch
additional information, Further Reading
agent tiers, Distribution: Agent Tiers–Delivery Guarantees Avro support, File Formats batch processing, Batching
component catalog, Component Catalog–Component Catalog example of, An Example–An Example fanning out, Fan Out–Delivery Guarantees
HDFS sinks and, The HDFS Sink–Delivery Guarantees installing, Installing Flume
integrating with applications, Integrating Flume with Applications sink groups, Sink Groups–Sink Groups
transactions and reliability, Transactions and Reliability–Batching
flume-ng command, Installing Flume, An Example
FlumeJava library, Crunch
Folding@home project, Volunteer Computing
FOREACH…GENERATE statement (Pig Latin), Statements, Statements, FOREACH… GENERATE
From class, Reading from a source FROM clause (Hive), Inner joins fs command, Statements
fs.datanode.dns.interface property, Hadoop Daemon Addresses and Ports fs.datanode.du.reserved property, Reserved storage space fs.defaultFS property about, The Command-Line Interface
finding namenode hostname, Starting and Stopping the Daemons
Hadoop modes and, Configuration
Hive and, Configuring Hive
Pig and, MapReduce mode
RPC servers, Hadoop Daemon Addresses and Ports setting filesystem, Testing the Driver specifying default filesystem, HDFS fs.file.impl property, LocalFileSystem
fs.trash.interval property, Trash
fsck tool, Blocks–Blocks, Filesystem check (fsck)–Finding the blocks for a file, Filesystem check (fsck)
FSDataInputStream class, FSDataInputStream–FSDataInputStream, Anatomy of a File Read
FSDataOutputStream class, FSDataOutputStream, Anatomy of a File Write, Coherency
Model
fsimage file, Namenode directory structure–The filesystem image and edit log, Safe Mode, Metadata backups
FsInput class, Avro Datafiles
FTPFileSystem class, Hadoop Filesystems
fully distributed mode (Hadoop), Fully Distributed Mode FuncSpec class, Leveraging types
functions (Crunch), Functions–Object reuse, Crunch Libraries–Crunch Libraries functions (Hive) about, Operators and Functions
UDF types, User-Defined Functions–User-Defined Functions writing UDAFs, Writing a UDAF–A more complex UDAF writing UDFs, Writing a UDF–Writing a UDF
functions (Pig Latin) built-in, Types, Functions–Functions types supported, Functions
user-defined, Other libraries, User-Defined Functions–Using a schema
functions (Spark), Transformations and Actions, Functions
FUSE (Filesystem in Userspace), FUSE
Future interface (Java), Asynchronous execution
G
GC_TIME_MILLIS counter, Task counters
GenericDatumWriter class, In-Memory Serialization and Deserialization
GenericOptionsParser class, GenericOptionsParser, Tool, and
ToolRunner–GenericOptionsParser, Tool, and ToolRunner, Usage
GenericRecord interface, In-Memory Serialization and Deserialization, Records and tuples
GenericWritable class, ObjectWritable and GenericWritable, Writable collections
Genome Reference Consortium (GRC), The Human Genome Project and Reference Genomes
Get class, Java
getACL operation (ZooKeeper), Operations getChildren operation (ZooKeeper), Operations getData operation (ZooKeeper), Operations GFS (Google), A Brief History of Apache Hadoop globbing operation, File patterns
Google GFS, A Brief History of Apache Hadoop Google Protocol Buffers, Serialization IDL graceful failover, Failover and fencing
Gradle build tool, Setting Up the Development Environment
Gray, Jim, Grid Computing
Gray, Jonathan, HBasics–Further Reading
GRC (Genome Reference Consortium), The Human Genome Project and Reference Genomes
Great Internet Mersenne Prime Search project, Volunteer Computing grid computing, Grid Computing
Gridmix benchmark suite, Other benchmarks
GROUP BY clause (Hive), An Example, Views
GROUP BY operator (Pig Latin), Comparison with Databases
GROUP statement (Pig Latin), Structure, Statements, Statements, GROUP grouping data, Grouping and Joining Data–GROUP groups (ZooKeeper) about, An Example
creating, Creating the Group–Creating the Group deleting, Deleting a Group
group membership, Group Membership in ZooKeeper joining, Joining a Group
listing members, Listing Members in a Group–ZooKeeper command-line tools Grunt shell, Running Pig Programs
gzip compression, Compression–Codecs, Native libraries
GzipCodec class, Codecs
H
Hadoop about, Beyond Batch
history of, A Brief History of Apache Hadoop–A Brief History of Apache Hadoop installing, Installing Apache Hadoop–Fully Distributed Mode
hadoop command
basic filesystem operations, Basic Filesystem Operations–Basic Filesystem Operations creating HAR files, Hadoop Filesystems distcp program and, Parallel Copying with distcp finding file checksum, Data Integrity in HDFS Hadoop Streaming and, Ruby launching JVM, A test run
retrieving job results, Retrieving the Results running miniclusters from, Testing the Driver
Hadoop Distributed Filesystem (see HDFS)
Hadoop Streaming about, Hadoop Streaming
MapReduce scripts and, MapReduce Scripts
Python example, Python
Ruby example, Ruby–Ruby hadoop-env.sh file, Hadoop Configuration, Metrics and JMX hadoop-metrics2.properties file, Hadoop Configuration, Metrics and JMX hadoop-policy.xml file, Hadoop Configuration, An example hadoop.http.staticuser.user property, Managing Configuration hadoop.rpc.protection property, Other Security Enhancements hadoop.security.authentication property, An example hadoop.security.authorization property, An example hadoop.ssl.enabled property, Other Security Enhancements
hadoop.user.group.static.mapping.overrides property, Managing Configuration
HADOOP_CLASSPATH environment variable, The client classpath, Launching a Job
HADOOP_CONF_DIR environment variable, Managing Configuration, Hadoop Configuration, MapReduce mode
HADOOP_HEAPSIZE environment variable, Memory heap size
HADOOP_HOME environment variable, MapReduce mode, Installing Hive, Installation
HADOOP_IDENT_STRING environment variable, System logfiles
HADOOP_LOG_DIR environment variable, Hadoop Logs, System logfiles
HADOOP_NAMENODE_OPTS environment variable, Memory heap size, Metrics and
JMX
HADOOP_OPTS environment variable, GenericOptionsParser, Tool, and ToolRunner
HADOOP_SSH_OPTS environment variable, SSH settings
HADOOP_USER_CLASSPATH_FIRST environment variable, Task classpath precedence
HADOOP_USER_NAME environment variable, Managing Configuration
Hammerbacher, Jeff, Hive
HAR files, Hadoop Filesystems
HarFileSystem class, Hadoop Filesystems
HashPartitioner class, Implementing a Custom Writable, The Default MapReduce Job, An example: Partitioning data
HBase about, Beyond Batch, The Design of HDFS, HBasics, HBase additional information, Further Reading building online query application about, Building an Online Query Application loading data, Loading Data–Bulk load
online queries, Online Queries–Observation queries schema design, Schema Design
client options, Clients–REST and Thrift common issues, Praxis–Counters
data model, Whirlwind Tour of the Data Model–Locking database input and output, Database Input (and Output) Hive and, Storage handlers
implementing, Implementation–HBase in operation installing, Installation–Test Drive
RDBMS comparison, HBase Versus RDBMS–HBase
test drive, Test Drive–Test Drive hbase.client.scanner.caching property, Java hbase.client.scanner.timeout.period property, Java hbase:meta table, HBase in operation
HBaseConfiguration class, Java
HBaseStorage function (Pig Latin), Functions
HCatalog (Hive), Using Hive tables with HCatalog
HDFS (Hadoop Distributed Filesystem)
Data Storage and Analysis, The Hadoop Distributed Filesystem audit logs, Hadoop Logs, Audit Logging basic concepts, Blocks–Failover and fencing
basic operations, Basic Filesystem Operations–Basic Filesystem Operations benchmarking, Benchmarking MapReduce with TeraSort cluster balancing, Keeping an HDFS Cluster Balanced cluster setup and installation, Creating Unix User Accounts cluster sizing, Master node scenarios
coherency model, Coherency Model–Consequences for application design command-line interface, The Command-Line Interface–Basic Filesystem Operations daemon properties, HDFS–HDFS
design overview, The Design of HDFS–The Design of HDFS file permissions, Basic Filesystem Operations
formatting filesystem, Formatting the HDFS Filesystem, Formatting the HDFS filesystem
HBase and, HDFS
high availability, HDFS High Availability–Failover and fencing Java interface, The Java Interface–Deleting Data
parallel copying with distcp, Parallel Copying with distcp–Keeping an HDFS Cluster Balanced
persistent data structures, Persistent Data Structures–Datanode directory structure reading files, Anatomy of a File Read–Anatomy of a File Read safe mode, Safe Mode–Entering and leaving safe mode scaling out data, Scaling Out–Data Flow
starting and stopping daemons, Starting and Stopping the Daemons tool support, dfsadmin–Balancer
upgrade considerations, HDFS data and metadata upgrades–Finalize the upgrade
(optional)
writing files, Anatomy of a File Write–Anatomy of a File Write
HDFS Federation, HDFS Federation
HDFS sinks
The HDFS Sink–The HDFS Sink fanning out, Fan Out–Delivery Guarantees file formats, File Formats
indexing events and, Delivery Guarantees
partitioning and interceptors, Partitioning and Interceptors
hdfs-site.xml file, Hadoop Configuration, Important Hadoop Daemon Properties, Commissioning new nodes hdfs.fileType property, File Formats hdfs.inUsePrefix property, The HDFS Sink
hdfs.path property, The HDFS Sink, Partitioning and Interceptors hdfs.proxyUser property, The HDFS Sink hdfs.rollInterval property, The HDFS Sink hdfs.rollSize property, The HDFS Sink
hdfs.useLocalTimeStamp property, Partitioning and Interceptors hdfs.writeFormat property, File Formats Hedwig system, BookKeeper and Hedwig help command, Statements herd effect, The herd effect
hexdump tool, The default storage format: Delimited text HFileOutputFormat2 class, Bulk load high availability
HDFS and, HDFS High Availability–Failover and fencing resource managers and, Resource Manager Failure
YARN and, YARN Compared to MapReduce 1 high-performance computing (HPC), Grid Computing history command, Statements
Hive
Hive additional information, Further Reading Avro support, Text and Binary File Formats
column-oriented format, Other File Formats and Column-Oriented Formats configuring Hive, Configuring Hive–Logging
database comparison, Comparison with Traditional Databases–SQL-on-Hadoop Alternatives
execution engines, Execution engines
HCatalog support, Using Hive tables with HCatalog installing, Installing Hive–The Hive Shell metastore, The Metastore–The Metastore
ORCFile and, Parquet
Parquet support, Text and Binary File Formats querying data, Querying Data–Views services supported, Hive Services–Hive clients
SQL dialect, HiveQL–Conversions
Sqoop exports and, Performing an Export
Squoop imports and, Imported Data and Hive–Imported Data and Hive tables about, Tables altering, Altering Tables
buckets and, Partitions and Buckets, Buckets–Buckets dropping, Dropping Tables
external tables, Managed Tables and External Tables–Managed Tables and External Tables
importing data, Importing Data–CREATE TABLE…AS SELECT
managed tables, Managed Tables and External Tables–Managed Tables and External Tables
partitions and, Partitions and Buckets–Partitions storage formats, Storage Formats–Storage handlers
version considerations, Installing Hive weather dataset example, An Example–An Example
Hive Web Interface (HWI), Hive Services
hive.execution.engine property, Execution engines hive.metastore.uris property, The Metastore hive.metastore.warehouse.dir property, The Metastore hive.server2.thrift.port property, Hive Services
HiveDriver class, Hive clients
HiveQL
about, The Hive Shell
data types, Data Types–Complex types operators and functions, Operators and Functions SQL comparison, HiveQL–HiveQL HiveServer2, Hive Services Hopper, Grace, Data!
HPC (high-performance computing), Grid Computing
HPROF profiling tool, The HPROF profiler
HTableDescriptor class, Java
HTTP REST API, HTTP
HTTP server properties, Hadoop Daemon Addresses and Ports
HttpFS proxy, HTTP, Parallel Copying with distcp
Human Genome Project, The Human Genome Project and Reference Genomes
HWI (Hive Web Interface), Hive Services
I
I/O (input/output) compression, Compression–Compressing map output data integrity, Data Integrity–ChecksumFileSystem
file-based data structures, File-Based Data Structures–Other File Formats and ColumnOriented Formats
serialization, Serialization–Serialization IDL
IDL (interface description language), Serialization IDL
ILLUSTRATE operator (Pig Latin), Generating Examples, Statements, Sorting Data
ImmutableBytesWritable class, MapReduce
Impala query engine, Application Lifespan, SQL-on-Hadoop Alternatives import process (Hive tables), Importing Data–CREATE TABLE…AS SELECT import process (Sqoop) consistency and, Imports and Consistency controlling, Controlling the Import direct-mode imports, Direct-Mode Imports
Hive and, Imported Data and Hive–Imported Data and Hive
importing large objects, Importing Large Objects–Importing Large Objects incremental imports, Incremental Imports
overview, Imports: A Deeper Look–Imports: A Deeper Look tool support, A Sample Import
working with imported data, Working with Imported Data–Working with Imported Data IMPORT statement (Pig Latin), Statements indexes (Hive), Updates, Transactions, and Indexes Infochimps.org, Data!
information commons, Data!
initLimit property, Configuration inner joins, Inner joins input formats binary input, Binary Input
database input, Database Input (and Output)
input splits and records, Data Flow, Compression and Input Splits, Input Splits and
Records–Processing a whole file as a record multiple inputs, Multiple Inputs text input, Text Input–XML
input splits about, Data Flow, Input Splits and Records–Input Splits and Records block and, TextInputFormat
CombineFileInputFormat class, Small files and CombineFileInputFormat compression and, Compression and Input Splits controlling split size, FileInputFormat input splits
FileInputFormat class, FileInputFormat–FileInputFormat input splits finding information on, File information in the mapper preventing, Preventing splitting
InputFormat class, Input Splits and Records, FileInputFormat, Imports: A Deeper Look InputSampler class, Total Sort
InputSplit class (Java), Input Splits and Records
InputStream class (Java), FSDataInputStream
INSERT INTO TABLE statement (Hive), Updates, Transactions, and Indexes, Inserts
INSERT OVERWRITE DIRECTORY statement (Hive), Managed Tables and External Tables
INSERT OVERWRITE TABLE statement (Hive), Buckets, Inserts interactive SQL, Beyond Batch
interface description language (IDL), Serialization IDL interfaces (Hadoop), Interfaces–FUSE
IntSumReducer class, MapReduce Library Classes
IntWritable class, Java MapReduce, The Writable Interface, Writable wrappers for Java primitives
InverseMapper class, MapReduce Library Classes
InvokeForDouble class, Dynamic invokers
InvokeForFloat class, Dynamic invokers
InvokeForInt class, Dynamic invokers
InvokeForLong class, Dynamic invokers InvokeForString class, Dynamic invokers
io.compression.codecs property, Inferring CompressionCodecs using CompressionCodecFactory
io.file.buffer.size property, Buffer size io.native.lib.available property, Native libraries io.serializations property, Serialization Frameworks
IOUtils class, Compressing and decompressing streams with CompressionCodec,
Processing a whole file as a record is null operator (Hive), Validation and nulls IsEmpty function (Pig Latin), Functions, Functions isro command (ZooKeeper), Installing and Running ZooKeeper iterative processing, Beyond Batch
J
jar service (Hive), Hive Services
Java Database Connectivity (JDBC), Imports: A Deeper Look, Exports: A Deeper Look Java language creating directories, Directories deleting data, Deleting Data environment variables, Java
Hadoop Streaming and, Ruby HBase and, Java–Java installing, Installing Java
Pig and, Running Pig Programs
querying FileSystem, File metadata: FileStatus–PathFilter reading data from Hadoop URL, Reading Data from a Hadoop URL reading data using FileSystem API, Reading Data Using the FileSystem API–FSDataInputStream secondary sort, Java code–Java code Spark example, A Java Example syslog file, Hadoop Logs
user-defined counters, User-Defined Java Counters–Retrieving counters WAR files, Packaging a Job
weather dataset example, Java MapReduce–A test run
Writable wrappers for Java primitives, Writable wrappers for Java primitives–Writable collections
writing data, Writing Data–FSDataOutputStream
Java Management Extensions (JMX), Metrics and JMX, Installing and Running ZooKeeper
Java Native Interface (JNI), C
Java Object Serialization, Serialization Frameworks
Java virtual machine (JVM), A test run, Remote Debugging, Task Failure java.library.path property, Native libraries java.util.concurrent package, Asynchronous execution
JavaPairRDD class, A Java Example
JavaRDD class, A Java Example
JavaRDDLike interface, A Java Example
JavaSerialization class, Serialization Frameworks
JAVA_HOME environment variable, Java, Installing and Running Pig, Installation
JBOD (just a bunch of disks), Cluster Specification
JDBC (Java Database Connectivity), Imports: A Deeper Look, Exports: A Deeper Look
JDBC drivers, Hive clients
JMX (Java Management Extensions), Metrics and JMX, Installing and Running ZooKeeper
JNI (Java Native Interface), C
Job class distributed cache options, The distributed cache API progress and status updates, Progress and Status Updates setting explicit JAR files, Packaging a Job setting input paths, FileInputFormat input paths
job counters, Built-in Counters, Job counters job history, The resource manager page job history logs (MapReduce), Hadoop Logs
job IDs, Launching a Job, The Task Execution Environment job JAR files about, Packaging a Job
client classpath, The client classpath packaging dependencies, Packaging dependencies task classpath, The task classpath
task classpath precedence, Task classpath precedence
job page (MapReduce), The MapReduce job page JobBuilder class, The Default MapReduce Job
JobClient class, Anatomy of a MapReduce Job Run
JobConf class, Compressing map output, Packaging a Job, The HPROF profiler JobControl class, JobControl jobs
anatomy of MapReduce job runs, Anatomy of a MapReduce Job Run–Job Completion anatomy of Spark job runs, Anatomy of a Spark Job Run–Task Execution completion process, Job Completion
DAG construction, DAG Construction–DAG Construction
debugging, Debugging a Job–Handling malformed data, Remote Debugging decomposing problems into, Decomposing a Problem into MapReduce Jobs–Decomposing a Problem into MapReduce Jobs
default MapReduce, The Default MapReduce Job–Keys and values in Streaming initialization process, Job Initialization launching, Launching a Job–Launching a Job logging, Hadoop Logs–Hadoop Logs
packaging, Packaging a Job–Task classpath precedence progress and status updates, Progress and Status Updates retrieving results, Retrieving the Results running as benchmarks, User Jobs
running locally, Running a Job in a Local Job Runner–Running a Job in a Local Job Runner
running Oozie workflow jobs, Running an Oozie workflow job scheduling, Job scheduler, Task Scheduling
Spark support, Spark Applications, Jobs, Stages, and Tasks submission process, Job Submission, Job Submission task execution, Task Execution
testing job drivers, Testing the Driver–Testing the Driver tuning, Tuning a Job–The HPROF profiler
viewing information about, The MapReduce Web UI–The MapReduce job page JobSubmitter class, Job Submission jobtrackers, YARN Compared to MapReduce 1
Join class, Crunch Libraries
JOIN clause (Hive), Inner joins
JOIN statement (Pig Latin), Statements, JOIN joins
about, Joins, Joins inner, Inner joins
map-side, Map-Side Joins–Map-Side Joins, Map joins outer, Outer joins
Pig operators and, Grouping and Joining Data–GROUP reduce-side, Reduce-Side Joins–Reduce-Side Joins semi, Semi joins
journal nodes, HDFS High Availability
JsonLoader function (Pig Latin), Functions
JsonStorage function (Pig Latin), Functions
JSP Expression Language, Defining an Oozie workflow JUnit framework, Setting Up the Development Environment just a bunch of disks (JBOD), Cluster Specification
JVM (Java virtual machine), A test run, Remote Debugging, Task Failure
JVMFLAGS environment variable, Resilience and Performance
K
KDC (Key Distribution Center), Kerberos and Hadoop kdestroy command, An example
Kellerman, Jim, Backdrop
Kerberos authentication, Kerberos and Hadoop–An example, ACLs
Key Distribution Center (KDC), Kerberos and Hadoop KeyValueTextInputFormat class, KeyValueTextInputFormat keywords (Pig Latin), Structure kill command, Statements
Kimball, Aaron, Sqoop–Further Reading kinit command, Kerberos and Hadoop klist command, An example ktutil command, Kerberos and Hadoop
L
LARGE_READ_OPS counter, Task counters
LATERAL VIEW statement (Hive), User-Defined Functions
LazyOutputFormat class, Lazy Output
leader election process, Implementation, A Lock Service leaderServes property, Consistency
LEFT OUTER JOIN statement (Hive), Outer joins LEFT SEMI JOIN statement (Hive), Semi joins libhdfs (C library), C
LIMIT statement (Pig Latin), Statements, Sorting Data Lin, Jimmy, MapReduce Workflows
line length, maximum, Controlling the maximum line length
LinuxContainerExecutor class, CPU settings in YARN and MapReduce, Other Security Enhancements
LinuxTaskController class, Other Security Enhancements list command (HBase), Test Drive listing files, Listing files
Llama project, Application Lifespan
load balancing, sink groups and, Sink Groups–Integrating Flume with Applications
LOAD DATA statement (Hive), An Example, Managed Tables and External Tables, Using a custom SerDe: RegexSerDe
load functions (Pig Latin), Functions, A Load UDF–Using a schema
LOAD statement (Pig Latin), Statements, Statements LoadFunc class, A Load UDF local mode (Hadoop), Standalone Mode local mode (Pig), Local mode
local mode (Spark), Executors and Cluster Managers
LocalFileSystem class, Hadoop Filesystems, LocalFileSystem locality constraints, Resource Requests
lock service (ZooKeeper), A Lock Service–Implementation locking HBase tables, Locking log aggregation, Hadoop Logs
log4j.properties file, Hadoop Configuration, Setting log levels logfiles
edit logs, The filesystem image and edit log–The filesystem image and edit log file-based data structures and, SequenceFile–The SequenceFile format
Flume support, An Example Hive support, Logging monitoring support, Logging
types supported, Hadoop Logs–Hadoop Logs, System logfiles
LongSumReducer class, MapReduce Library Classes
LongWritable class, Java MapReduce, Writable wrappers for Java primitives, MapReduce
Types
ls command, Structure, Statements
LZ4 compression, Compression–Codecs, Native libraries
Lz4Codec class, Codecs
LZO compression, Compression–Codecs, Native libraries LzoCodec class, Codecs lzop tool, Codecs
LzopCodec class, Codecs
M
macros (Pig Latin), Macros–Macros MailTrust, Querying All Your Data maintenance (see system administration)
managed tables (Hive), Managed Tables and External Tables–Managed Tables and External Tables
MAP clause (Hive), MapReduce Scripts map functions (MapReduce)
about, Map and Reduce
compressing output, Compressing map output data flow tasks, Data Flow–Specifying a combiner function general form, MapReduce Types
Hadoop Streaming, Hadoop Streaming
HDFS blocks and, Blocks Java example, Java MapReduce joining data, Map-Side Joins–Map-Side Joins progress and status updates, Progress and Status Updates shuffle and sort, The Map Side–The Map Side Spark and, DAG Construction task assignments, Task Assignment task execution, Task Execution task failures, Task Failure testing with MRUnit, Mapper–Mapper tuning checklist, Tuning a Job tuning properties, Configuration Tuning
MapDriver class, Mapper
MapFile class, MapFile
MapFileOutputFormat class, MapFileOutputFormat
MapFn class, parallelDo(), combineValues()
Mapper interface about, Mapper–Mapper
finding information on input splits, File information in the mapper task execution, The Task Execution Environment type parameters, MapReduce Types Mapred class, Crunch Libraries mapred-env.sh file, Hadoop Configuration mapred-site.xml file, Hadoop Configuration
mapred.child.java.opts property, Remote Debugging, Configuration Tuning, Memory settings in YARN and MapReduce
mapred.combiner.class property, MapReduce Types mapred.input.format.class property, MapReduce Types mapred.job.tracker property, Running a Job in a Local Job Runner mapred.map.runner.class property, MapReduce Types mapred.mapoutput.key.class property, MapReduce Types mapred.mapoutput.value.class property, MapReduce Types mapred.mapper.class property, MapReduce Types mapred.output.format.class property, MapReduce Types mapred.output.key.class property, MapReduce Types mapred.output.key.comparator.class property, MapReduce Types mapred.output.value.class property, MapReduce Types mapred.output.value.groupfn.class property, MapReduce Types mapred.partitioner.class property, MapReduce Types mapred.reducer.class property, MapReduce Types
MapReduce about, Data Storage and Analysis, MapReduce, MapReduce Workflows anatomy of job runs, Anatomy of a MapReduce Job Run–Job Completion Avro support, Avro MapReduce–Sorting Using Avro MapReduce batch processing, Beyond Batch
benchmarking with TeraSort, Benchmarking MapReduce with TeraSort cluster setup and installation, Creating Unix User Accounts
compression and, Compression and Input Splits–Compressing map output counters, Counters–User-Defined Streaming Counters Crunch and, Crunch
daemon properties, Memory settings in YARN and MapReduce–CPU settings in YARN and MapReduce
decomposing problems into jobs, Decomposing a Problem into MapReduce Jobs–Decomposing a Problem into MapReduce Jobs
default job, The Default MapReduce Job–Keys and values in Streaming developing applications
about, Developing a MapReduce Application
Configuration API, The Configuration API–Variable Expansion
running locally on test data, Running Locally on Test Data–Testing the Driver running on clusters, Running on a Cluster–Remote Debugging setting up development environment, Setting Up the Development Environment–GenericOptionsParser, Tool, and ToolRunner tuning jobs, Tuning a Job–The HPROF profiler
workflows, MapReduce Workflows–Running an Oozie workflow job writing unit tests, Writing a Unit Test with MRUnit–Reducer
failure considerations, Failures–Resource Manager Failure
Hadoop Streaming, Hadoop Streaming–Python
HBase and, MapReduce–MapReduce Hive and, Execution engines
input formats, Input Formats–Database Input (and Output) joining data, Joins–Reduce-Side Joins
library classes supported, MapReduce Library Classes
old and new API comparison, The Old and New Java MapReduce APIs–The Old and New Java MapReduce APIs old API signatures, MapReduce Types output formats, Output Formats–Database Output Parquet support, Parquet MapReduce–Parquet MapReduce progress reporting in, Progress and Status Updates querying data, Querying All Your Data, MapReduce Scripts resource requests, Resource Requests
shuffle and sort, Shuffle and Sort–Configuration Tuning
side data distribution, Side Data Distribution–The distributed cache API
sorting data, Sorting–Streaming, Sorting Using Avro MapReduce–Sorting Using Avro MapReduce
Spark and, Transformations and Actions
Sqoop support, A Sample Import, Imports: A Deeper Look, Exports: A Deeper Look starting and stopping daemons, Starting and Stopping the Daemons
system comparison, Relational Database Management Systems–Volunteer Computing task execution, Task Execution, Task Execution–Task side-effect files
types supported, MapReduce Types–The Default MapReduce Job
weather dataset example, A Weather Dataset–Running a Distributed MapReduce Job
YARN comparison, YARN Compared to MapReduce 1–YARN Compared to MapReduce 1
Mapreduce class, Crunch Libraries
MapReduce mode (Pig), MapReduce mode, Parallelism MAPREDUCE statement (Pig Latin), Statements
mapreduce.am.max-attempts property, Application Master Failure
mapreduce.client.progressmonitor.pollinterval property, Progress and Status Updates mapreduce.client.submit.file.replication property, Job Submission mapreduce.cluster.acls.enabled property, Other Security Enhancements mapreduce.cluster.local.dir property, Remote Debugging, The Map Side
mapreduce.framework.name property, Running a Job in a Local Job Runner, Testing the Driver, MapReduce mode, Configuration
mapreduce.input.fileinputformat.input.dir.recursive property, FileInputFormat input paths mapreduce.input.fileinputformat.inputdir property, FileInputFormat input paths mapreduce.input.fileinputformat.split.maxsize property, FileInputFormat input splits mapreduce.input.fileinputformat.split.minsize property, FileInputFormat input splits
mapreduce.input.keyvaluelinerecordreader.key.value.separator property, KeyValueTextInputFormat
mapreduce.input.lineinputformat.linespermap property, NLineInputFormat
mapreduce.input.linerecordreader.line.maxlength property, Controlling the maximum line length
mapreduce.input.pathFilter.class property, FileInputFormat input paths mapreduce.job.acl-modify-job property, Other Security Enhancements mapreduce.job.acl-view-job property, Other Security Enhancements mapreduce.job.combine.class property, MapReduce Types mapreduce.job.end-notification.url property, Job Completion mapreduce.job.hdfs-servers property, Delegation Tokens mapreduce.job.id property, The Task Execution Environment mapreduce.job.inputformat.class property, MapReduce Types mapreduce.job.map.class property, MapReduce Types
mapreduce.job.maxtaskfailures.per.tracker property, Node Manager Failure mapreduce.job.output.group.comparator.class property, MapReduce Types mapreduce.job.output.key.class property, MapReduce Types
mapreduce.job.output.key.comparator.class property, MapReduce Types, Partial Sort mapreduce.job.output.value.class property, MapReduce Types mapreduce.job.outputformat.class property, MapReduce Types mapreduce.job.partitioner.class property, MapReduce Types mapreduce.job.queuename property, Queue placement mapreduce.job.reduce.class property, MapReduce Types
mapreduce.job.reduce.slowstart.completedmaps property, Reduce slow start mapreduce.job.reduces property, Job Initialization mapreduce.job.ubertask.enable property, Job Initialization mapreduce.job.ubertask.maxbytes property, Job Initialization mapreduce.job.ubertask.maxmaps property, Job Initialization mapreduce.job.ubertask.maxreduces property, Job Initialization mapreduce.job.user.classpath.first property, Task classpath precedence mapreduce.jobhistory.address property, Hadoop Daemon Addresses and Ports mapreduce.jobhistory.bind-host property, Hadoop Daemon Addresses and Ports mapreduce.jobhistory.webapp.address property, Hadoop Daemon Addresses and Ports mapreduce.map.combine.minspills property, The Map Side, Configuration Tuning mapreduce.map.cpu.vcores property, Task Assignment, CPU settings in YARN and MapReduce
mapreduce.map.failures.maxpercent property, Task Failure mapreduce.map.input.file property, File information in the mapper mapreduce.map.input.length property, File information in the mapper mapreduce.map.input.start property, File information in the mapper mapreduce.map.java.opts property, Memory settings in YARN and MapReduce mapreduce.map.log.level property, Hadoop Logs mapreduce.map.maxattempts property, Task Failure
mapreduce.map.memory.mb property, Task Assignment, Memory settings in YARN and MapReduce
mapreduce.map.output.compress property, Compressing map output, The Map Side, Configuration Tuning
mapreduce.map.output.compress.codec property, Compressing map output, The Map Side, Configuration Tuning
mapreduce.map.output.key.class property, MapReduce Types mapreduce.map.output.value.class property, MapReduce Types
mapreduce.map.sort.spill.percent property, The Map Side, Configuration Tuning mapreduce.map.speculative property, Speculative Execution
mapreduce.mapper.multithreadedmapper.threads property, Input Splits and Records mapreduce.output.fileoutputformat.compress property, Using Compression in MapReduce, Parquet Configuration
mapreduce.output.fileoutputformat.compress.codec property, Using Compression in MapReduce
mapreduce.output.fileoutputformat.compress.type property, Using Compression in MapReduce
mapreduce.output.textoutputformat.separator property, Text Output
mapreduce.reduce.cpu.vcores property, Task Assignment, CPU settings in YARN and MapReduce
mapreduce.reduce.failures.maxpercent property, Task Failure
mapreduce.reduce.input.buffer.percent property, Configuration Tuning, Configuration Tuning
mapreduce.reduce.java.opts property, Memory settings in YARN and MapReduce mapreduce.reduce.log.level property, Hadoop Logs mapreduce.reduce.maxattempts property, Task Failure
mapreduce.reduce.memory.mb property, Task Assignment, Memory settings in YARN and MapReduce
mapreduce.reduce.merge.inmem.threshold property, The Reduce Side, Configuration Tuning, Configuration Tuning
mapreduce.reduce.shuffle.input.buffer.percent property, The Reduce Side, Configuration Tuning
mapreduce.reduce.shuffle.maxfetchfailures property, Configuration Tuning
mapreduce.reduce.shuffle.merge.percent property, The Reduce Side, Configuration Tuning mapreduce.reduce.shuffle.parallelcopies property, The Reduce Side, Configuration Tuning mapreduce.reduce.speculative property, Speculative Execution
mapreduce.shuffle.max.threads property, The Map Side, Configuration Tuning mapreduce.shuffle.port property, Hadoop Daemon Addresses and Ports mapreduce.shuffle.ssl.enabled property, Other Security Enhancements mapreduce.task.attempt.id property, The Task Execution Environment mapreduce.task.files.preserve.failedtasks property, Remote Debugging mapreduce.task.files.preserve.filepattern property, Remote Debugging mapreduce.task.id property, The Task Execution Environment
mapreduce.task.io.sort.factor property, The Map Side, The Reduce Side, Configuration Tuning
mapreduce.task.io.sort.mb property, The Map Side, Configuration Tuning mapreduce.task.ismap property, The Task Execution Environment mapreduce.task.output.dir property, Task side-effect files mapreduce.task.partition property, The Task Execution Environment mapreduce.task.profile property, The HPROF profiler mapreduce.task.profile.maps property, The HPROF profiler mapreduce.task.profile.reduces property, The HPROF profiler mapreduce.task.timeout property, Task Failure mapreduce.task.userlog.limit.kb property, Hadoop Logs
MapWritable class, Writable collections
MAP_INPUT_RECORDS counter, Task counters
MAP_OUTPUT_BYTES counter, Task counters
MAP_OUTPUT_MATERIALIZED_BYTES counter, Task counters MAP_OUTPUT_RECORDS counter, Task counters mashups, Data!
Massie, Matt, Biological Data Science: Saving Lives with Software master nodes (HBase), Implementation
master−worker pattern (namenodes), Namenodes and Datanodes materialization process (Crunch), Materialization–PObject
Maven POM (Project Object Model), Setting Up the Development Environment–Setting Up the Development Environment, Packaging a Job, The Specific API
MAX function (Pig Latin), Validation and nulls, Functions
MB_MILLIS_MAPS counter, Job counters MB_MILLIS_REDUCES counter, Job counters memory management
buffering writes, The Map Side
container virtual memory constraints, Memory settings in YARN and MapReduce daemons, Memory heap size memory heap size, Memory heap size
namenodes, Master node scenarios, Memory heap size Spark and, Spark
task assignments, Task Assignment
MemPipeline class, union()
MERGED_MAP_OUTPUTS counter, Task counters Message Passing Interface (MPI), Grid Computing metadata backups of, Metadata backups block sizes and, Blocks
filesystems and, The filesystem image and edit log–The filesystem image and edit log Hive metastore, Installing Hive, Hive Services, The Metastore–The Metastore namenode memory requirements, The Design of HDFS Parquet considerations, Parquet File Format
querying, File metadata: FileStatus–File metadata: FileStatus
upgrade considerations, HDFS data and metadata upgrades–Finalize the upgrade
(optional) metastore (Hive), Installing Hive, Hive Services, The Metastore–The Metastore METASTORE_PORT environment variable, Hive Services metrics counters and, Metrics and JMX HBase and, Metrics
JMX and, Metrics and JMX
Microsoft Research MyLifeBits project, Data!
MILLIS_MAPS counter, Job counters
MILLIS_REDUCES counter, Job counters MIN function (Pig Latin), Functions miniclusters, testing in, Testing the Driver minimal replication condition, Safe Mode
MIP (Message Passing Interface), Grid Computing
mkdir command, Statements
mntr command (ZooKeeper), Installing and Running ZooKeeper monitoring clusters about, Monitoring logging support, Logging metrics and JMX, Metrics and JMX
MorphlineSolrSink class, Delivery Guarantees
MRBench benchmark, Other benchmarks
MRUnit library about, Setting Up the Development Environment, Writing a Unit Test with MRUnit testing map functions, Mapper–Mapper testing reduce functions, Reducer
multiple files input formats, Multiple Inputs
MultipleOutputs class, MultipleOutputs–MultipleOutputs output formats, Multiple Outputs–MultipleOutputs
partitioning data, An example: Partitioning data–An example: Partitioning data
MultipleInputs class, Multiple Inputs, Reduce-Side Joins
MultipleOutputFormat class, Multiple Outputs
MultipleOutputs class, MultipleOutputs–MultipleOutputs multiplexing selectors, Replicating and Multiplexing Selectors multiquery execution, Statements multitable insert, Multitable insert
MultithreadedMapper class, Input Splits and Records, MapReduce Library Classes MultithreadedMapRunner class, MapReduce Library Classes mv command, Statements
MyLifeBits project, Data!
MySQL
creating database schemas, A Sample Import
Hive and, The Metastore HiveQL and, The Hive Shell
installing and configuring, A Sample Import populating database, A Sample Import
mysqlimport utility, Exports: A Deeper Look
N
namenodes about, A Brief History of Apache Hadoop, Namenodes and Datanodes block caching, Block Caching
checkpointing process, The filesystem image and edit log cluster setup and installation, Formatting the HDFS Filesystem cluster sizing, Master node scenarios
commissioning nodes, Commissioning new nodes–Commissioning new nodes data integrity and, Data Integrity in HDFS DataStreamer class and, Anatomy of a File Write
decommissioning nodes, Decommissioning old nodes–Decommissioning old nodes DFSInputStream class and, Anatomy of a File Read
directory structure, Namenode directory structure–Namenode directory structure failover controllers and, Failover and fencing filesystem metadata and, The Design of HDFS HDFS federation, HDFS Federation
memory considerations, Master node scenarios, Memory heap size replica placement, Anatomy of a File Write safe mode, Safe Mode–Entering and leaving safe mode
secondary, Namenodes and Datanodes, Starting and Stopping the Daemons, Secondary namenode directory structure
single points of failure, HDFS High Availability
starting, Starting and Stopping the Daemons, The filesystem image and edit log
namespaceID identifier, Namenode directory structure
National Climatic Data Center (NCDC)
data format, Data Format encapsulating parsing logic, Mapper multiple inputs, Multiple Inputs
preparing weather datafiles, Preparing the NCDC Weather Data–Preparing the NCDC Weather Data
NativeAzureFileSystem class, Hadoop Filesystems
NCDC (National Climatic Data Center) data format, Data Format encapsulating parsing logic, Mapper multiple inputs, Multiple Inputs
preparing weather datafiles, Preparing the NCDC Weather Data–Preparing the NCDC Weather Data
NDFS (Nutch Distributed Filesystem), A Brief History of Apache Hadoop nested encoding, Nested Encoding
net.topology.node.switch.mapping.impl property, Rack awareness net.topology.script.file.name property, Rack awareness
network topology, Anatomy of a File Read, Anatomy of a File Write, Network Topology–Rack awareness
NFS gateway, NFS
NLineInputFormat class, Task side-effect files, NLineInputFormat NNBench benchmark, Other benchmarks node managers
about, Anatomy of a YARN Application Run blacklisting, Node Manager Failure
commissioning nodes, Commissioning new nodes–Commissioning new nodes decommissioning nodes, Decommissioning old nodes–Decommissioning old nodes failure considerations, Node Manager Failure heartbeat requests, Delay Scheduling job initialization process, Job Initialization resource manager failure, Resource Manager Failure starting, Starting and Stopping the Daemons streaming tasks, Streaming task execution, Task Execution task failure, Task Failure
tasktrackers and, YARN Compared to MapReduce 1
normalization (data), Relational Database Management Systems
NullOutputFormat class, Text Output
NullWritable class, Mutability, NullWritable, Text Output
NUM_FAILED_MAPS counter, Job counters
NUM_FAILED_REDUCES counter, Job counters
NUM_FAILED_UBERTASKS counter, Job counters
NUM_KILLED_MAPS counter, Job counters
NUM_KILLED_REDUCES counter, Job counters
NUM_UBER_SUBMAPS counter, Job counters
NUM_UBER_SUBREDUCES counter, Job counters
Nutch Distributed Filesystem (NDFS), A Brief History of Apache Hadoop
Nutch search engine, A Brief History of Apache Hadoop–A Brief History of Apache Hadoop
O
Object class (Java), Implementing a Custom Writable object properties, printing, GenericOptionsParser, Tool, and ToolRunner–GenericOptionsParser, Tool, and ToolRunner
ObjectWritable class, ObjectWritable and GenericWritable ODBC drivers, Hive clients
oozie command-line tool, Running an Oozie workflow job
oozie.wf.application.path property, Running an Oozie workflow job OOZIE_URL environment variable, Running an Oozie workflow job operations (ZooKeeper) exceptions supported, The Resilient ZooKeeper Application–A reliable configuration service, Recoverable exceptions language bindings, APIs multiupdate, Multiupdate watch triggers, Watch triggers znode supported, Operations
operators (HiveQL), Operators and Functions operators (Pig) combining and splitting data, Combining and Splitting Data filtering data, Filtering Data–STREAM
grouping and joining data, Grouping and Joining Data–GROUP loading and storing data, Loading and Storing Data sorting data, Sorting Data
Optimized Record Columnar File (ORCFile), Parquet, Binary storage formats: Sequence files, Avro datafiles, Parquet files, RCFiles, and ORCFiles
ORCFile (Optimized Record Columnar File), Parquet, Binary storage formats: Sequence files, Avro datafiles, Parquet files, RCFiles, and ORCFiles
OrcStorage function (Pig Latin), Functions
ORDER BY clause (Hive), Sorting and Aggregating ORDER statement (Pig Latin), Statements, Sorting Data org.apache.avro.mapreduce package, Avro MapReduce org.apache.crunch.io package, Reading from a source org.apache.crunch.lib package, Crunch Libraries org.apache.flume.serialization package, File Formats org.apache.hadoop.classification package, Upgrades org.apache.hadoop.conf package, The Configuration API org.apache.hadoop.hbase package, Java
org.apache.hadoop.hbase.mapreduce package, MapReduce org.apache.hadoop.hbase.util package, Java
org.apache.hadoop.io package, Java MapReduce, Writable Classes org.apache.hadoop.io.serializer package, Serialization Frameworks org.apache.hadoop.mapreduce package, Input Splits and Records org.apache.hadoop.mapreduce.jobcontrol package, JobControl org.apache.hadoop.mapreduce.join package, Map-Side Joins org.apache.hadoop.streaming.mapreduce package, XML org.apache.pig.builtin package, A Filter UDF
org.apache.spark.rdd package, Transformations and Actions OTHER_LOCAL_MAPS counter, Job counters
outer joins, Outer joins output formats binary output, SequenceFileOutputFormat database output, Database Input (and Output) lazy output, Lazy Output
multiple outputs, Multiple Outputs–MultipleOutputs text output, Text Output
OutputCollector interface, Task side-effect files
OutputCommitter class, Job Initialization, Task Execution, Output Committers–Task sideeffect files
OutputFormat interface, Output Committers, Output Formats–Database Output
OVERWRITE keyword (Hive), An Example
OVERWRITE write mode, Existing outputs
O’Malley, Owen, A Brief History of Apache Hadoop
P
packaging jobs about, Packaging a Job
client classpath, The client classpath
packaging dependencies, Packaging dependencies task classpath, The task classpath
task classpath precedence, Task classpath precedence
packaging Oozie workflow applications, Packaging and deploying an Oozie workflow application
PageRank algorithm, Iterative Algorithms
Pair class, parallelDo(), combineValues()
PairRDDFunctions class, A Scala Standalone Application
PARALLEL keyword (Pig Latin), FOREACH…GENERATE, Parallelism parallel processing, Parallel Copying with distcp–Keeping an HDFS Cluster Balanced ParallelDo fusion, Inspecting a Crunch Plan
parameter substitution (Pig), Parameter Substitution–Parameter substitution processing
Parquet about, Other File Formats and Column-Oriented Formats, Parquet Avro and, Avro, Protocol Buffers, and Thrift–Projection and read schemas binary storage format and, Binary storage formats: Sequence files, Avro datafiles,
Parquet files, RCFiles, and ORCFiles configuring, Parquet Configuration data model, Data Model–Nested Encoding file format, Parquet File Format–Parquet File Format
Hive support, Text and Binary File Formats
MapReduce support, Parquet MapReduce–Parquet MapReduce nested encoding, Nested Encoding
Protocol Buffers and, Avro, Protocol Buffers, and Thrift–Projection and read schemas
Sqoop support, Text and Binary File Formats
Thrift and, Avro, Protocol Buffers, and Thrift–Projection and read schemas tool support, Parquet
writing and reading files, Writing and Reading Parquet Files–Projection and read schemas
parquet.block.size property, Parquet Configuration, Parquet MapReduce parquet.compression property, Parquet Configuration parquet.dictionary.page.size property, Parquet Configuration parquet.enable.dictionary property, Parquet Configuration parquet.example.data package, Writing and Reading Parquet Files parquet.example.data.simple package, Writing and Reading Parquet Files parquet.page.size property, Parquet Configuration
ParquetLoader function (Pig Latin), Functions
ParquetReader class, Writing and Reading Parquet Files
ParquetStorer function (Pig Latin), Functions
ParquetWriter class, Writing and Reading Parquet Files
partial sort, Partial Sort–Partial Sort
PARTITION clause (Hive), Inserts
PARTITIONED BY clause (Hive), Partitions partitioned data about, Relational Database Management Systems HDFS sinks and, Partitioning and Interceptors Hive tables and, Partitions and Buckets–Partitions
weather dataset example, An example: Partitioning data–An example: Partitioning data
Partitioner interface, MapReduce Types, Reduce-Side Joins
Path class, Reading Data Using the FileSystem API, Writing Data
PATH environment variable, HDFS data and metadata upgrades
PathFilter interface, Listing files–PathFilter
Paxos algorithm, Implementation
PCollection interface about, An Example asCollection() method, PObject
checkpointing pipelines, Checkpointing a Pipeline materialize() method, Materialization–PObject
parallelDo() method, An Example, parallelDo()–parallelDo(), Inspecting a Crunch Plan pipeline execution, Pipeline Execution reading files, Reading from a source types supported, Types–Records and tuples union() method, union() writing files, Writing to a target
permissions
ACL, ACLs
HDFS considerations, Basic Filesystem Operations storing, Blocks
persistence, RDD, Persistence–Persistence levels persistent data structures, Persistent Data Structures persistent znodes, Ephemeral znodes
PGroupedTable interface
about, An Example, groupByKey()
combineValues() method, combineValues()–combineValues(), Object reuse mapValues() method, Object reuse
PHYSICAL_MEMORY_BYTES counter, Task counters, Memory settings in YARN and MapReduce
Pig about, Pig
additional information, Further Reading anonymous relations and, Anonymous Relations
comparison with databases, Comparison with Databases–Comparison with Databases Crunch and, Crunch
data processing operators, Loading and Storing Data–Combining and Splitting Data execution types, Execution Types–MapReduce mode
installing and running, Installing and Running Pig–Pig Latin Editors parallelism and, Parallelism
parameter substitution and, Parameter Substitution–Parameter substitution processing practical techniques, Pig in Practice–Parameter substitution processing sorting data, Total Sort
user-defined functions, User-Defined Functions–Using a schema weather dataset example, An Example–Generating Examples
Pig Latin about, Pig, Pig Latin built-in types, Types–Types commands supported, Statements editor support, Pig Latin Editors expressions, Expressions–Expressions functions, Types, Functions–Other libraries macros, Macros–Macros schemas, Schemas–Schema merging statements, Statements–Statements structure, Structure
pig.auto.local.enabled property, MapReduce mode pig.auto.local.input.maxbytes, MapReduce mode PigRunner class, Running Pig Programs
PigServer class, Running Pig Programs
PigStorage function (Pig Latin), Functions
PIG_CONF_DIR environment variable, MapReduce mode pipeline execution (Crunch)
about, Pipeline Execution
checkpointing pipelines, Checkpointing a Pipeline
inspecting plans, Inspecting a Crunch Plan–Inspecting a Crunch Plan iterative algorithms, Iterative Algorithms–Iterative Algorithms running pipelines, Running a Pipeline–Debugging stopping pipelines, Stopping a Pipeline
Pipeline interface done() method, Stopping a Pipeline enableDebug() method, Debugging read() method, Reading from a source readTextFile() method, An Example
run() method, Running a Pipeline–Asynchronous execution runAsync() method, Asynchronous execution
PipelineExecution interface, Stopping a Pipeline
PipelineResult class, An Example, Running a Pipeline
PObject interface, PObject, Iterative Algorithms PositionedReadable interface, FSDataInputStream preemption, Preemption
PrimitiveEvalFunc class, An Eval UDF
printing object properties, GenericOptionsParser, Tool, and ToolRunner–GenericOptionsParser, Tool, and ToolRunner profiling tasks, Profiling Tasks–The HPROF profiler progress, tracking for tasks, Progress and Status Updates Progressable interface, Writing Data properties
daemon, Important Hadoop Daemon Properties–CPU settings in YARN and MapReduce
map-side tuning, Configuration Tuning
printing for objects, GenericOptionsParser, Tool, and ToolRunner–GenericOptionsParser, Tool, and ToolRunner reduce-side tuning, Configuration Tuning znodes, Data Model–Watches
Protocol Buffers, Avro, Protocol Buffers, and Thrift–Projection and read schemas ProtoParquetWriter class, Avro, Protocol Buffers, and Thrift psdsh shell tool, Configuration Management
pseudodistributed mode (Hadoop), Pseudodistributed Mode–Creating a user directory
PTable interface about, An Example asMap() method, PObject creating instance, parallelDo()
finding set of unique values for keys, Object reuse groupByKey() method, groupByKey() materializeToMap() method, Materialization reading text files, Reading from a source
PTables class, Crunch Libraries
PTableType interface, An Example
PType interface, parallelDo(), Types–Records and tuples, Object reuse Public Data Sets, Data! pwd command, Statements PySpark API, A Python Example pyspark command, A Python Example
Python language Avro and, Python API
incrementing counters, User-Defined Streaming Counters querying data, MapReduce Scripts Spark example, A Python Example weather dataset example, Python
Q
QJM (quorum journal manager), HDFS High Availability querying data about, Querying All Your Data aggregating data, Sorting and Aggregating batch processing, Beyond Batch
FileStatus class, File metadata: FileStatus–File metadata: FileStatus
FileSystem class, File metadata: FileStatus–PathFilter
HBase online query application, Building an Online Query Application–Observation queries
joining data, Joins–Map joins
MapReduce scripts, MapReduce Scripts sorting data, Sorting and Aggregating subqueries, Subqueries views, Views
queue elasticity, Capacity Scheduler Configuration queues
Capacity Scheduler, Capacity Scheduler Configuration–Queue placement
Fair Scheduler, Fair Scheduler Configuration–Preemption quit command, Statements
quorum journal manager (QJM), HDFS High Availability
R
r (read) permission, Basic Filesystem Operations rack local tasks, Task Assignment
rack topology, Rack awareness–Rack awareness Rackspace MailTrust, Querying All Your Data
RACK_LOCAL_MAPS counter, Job counters
RAID (redundant array of independent disks), Cluster Specification Rajaraman, Anand, Data!
RANK statement (Pig Latin), Statements
RawComparator interface, WritableComparable and comparators, Implementing a RawComparator for speed, Partial Sort
RawLocalFileSystem class, Hadoop Filesystems, LocalFileSystem
RDBMSs (Relational Database Management Systems)
about, Relational Database Management Systems–Relational Database Management Systems
HBase comparison, HBase Versus RDBMS–HBase
Hive metadata and, Tables
Pig comparison, Comparison with Databases
RDD class filter() method, An Example map() method, An Example
RDDs (Resilient Distributed Datasets) about, An Example, Resilient Distributed Datasets creating, Creation Java and, A Java Example
operations on, Transformations and Actions–Aggregation transformations persistence and, Persistence–Persistence levels serialization, Serialization
read (r) permission, Basic Filesystem Operations READ permission (ACL), ACLs reading data
Crunch support, Reading from a source
FileSystem class and, Reading Data Using the FileSystem API–FSDataInputStream,
Anatomy of a File Read
from Hadoop URL, Reading Data from a Hadoop URL
HDFS data flow, Anatomy of a File Read–Anatomy of a File Read
Parquet and, Writing and Reading Parquet Files–Projection and read schemas SequenceFile class, Reading a SequenceFile–Reading a SequenceFile short-circuiting local reads, Short-circuit local reads
ReadSupport class, Writing and Reading Parquet Files
READ_OPS counter, Task counters
RecordReader class, Input Splits and Records, Processing a whole file as a record records, processing files as, Processing a whole file as a record–Processing a whole file as a record
REDUCE clause (Hive), MapReduce Scripts reduce functions (MapReduce)
about, Map and Reduce
data flow tasks, Data Flow–Specifying a combiner function general form, MapReduce Types
Hadoop Streaming, Hadoop Streaming Java example, Java MapReduce
joining data, Reduce-Side Joins–Reduce-Side Joins progress and status updates, Progress and Status Updates shuffle and sort, The Reduce Side–The Reduce Side Spark and, DAG Construction task assignments, Task Assignment task execution, Task Execution task failures, Task Failure testing with MRUnit, Reducer tuning checklist, Tuning a Job tuning properties, Configuration Tuning
ReduceDriver class, Reducer
Reducer interface, The Task Execution Environment, MapReduce Types
REDUCE_INPUT_GROUPS counter, Task counters
REDUCE_INPUT_RECORDS counter, Task counters
REDUCE_OUTPUT_RECORDS counter, Task counters REDUCE_SHUFFLE_BYTES counter, Task counters
redundant array of independent disks (RAID), Cluster Specification reference genomes, The Human Genome Project and Reference Genomes
ReflectionUtils class, Compressing and decompressing streams with CompressionCodec, Reading a SequenceFile
RegexMapper class, MapReduce Library Classes RegexSerDe class, Using a custom SerDe: RegexSerDe regionservers (HBase), Implementation REGISTER statement (Pig Latin), Statements regular expressions, Using a custom SerDe: RegexSerDe Relational Database Management Systems (see RDBMSs) remote debugging, Remote Debugging remote procedure calls (RPCs), Serialization
replicated mode (ZooKeeper), Implementation, Configuration Reporter interface, Progress and Status Updates reserved storage space, Reserved storage space Resilient Distributed Datasets (see RDDs) resource manager page, The resource manager page resource managers about, Anatomy of a YARN Application Run application master failure, Application Master Failure cluster sizing, Master node scenarios
commissioning nodes, Commissioning new nodes–Commissioning new nodes decommissioning nodes, Decommissioning old nodes–Decommissioning old nodes failure considerations, Resource Manager Failure heartbeat requests, Delay Scheduling job initialization process, Job Initialization job submission process, Job Submission jobtrackers and, YARN Compared to MapReduce 1 node manager failure, Node Manager Failure progress and status updates, Progress and Status Updates starting, Starting and Stopping the Daemons task assignments, Task Assignment task execution, Task Execution thread dumps, Getting stack traces
resource requests, Resource Requests
REST, HBase and, REST and Thrift
Result class, Java
ResultScanner interface, Java
ResultSet interface, Imports: A Deeper Look rg.apache.hadoop.hbase.client package, Java rm command, Statements rmf command, Statements
ROW FORMAT clause (Hive), An Example, Using a custom SerDe: RegexSerDe, UserDefined Functions
RowCounter class, MapReduce
RPC server properties, Hadoop Daemon Addresses and Ports
RpcClient class (Java), Integrating Flume with Applications
RPCs (remote procedure calls), Serialization Ruby language, Ruby–Ruby run command, Statements, Statements
Runnable interface (Java), Building YARN Applications ruok command (ZooKeeper), Installing and Running ZooKeeper
S
S3AFileSystem class, Hadoop Filesystems safe mode, Safe Mode–Entering and leaving safe mode
Sammer, Eric, Setting Up a Hadoop Cluster Sample class, Crunch Libraries
SAMPLE statement (Pig Latin), Statements
Scala application example, A Scala Standalone Application–A Scala Standalone
Application scaling out (data) about, Scaling Out
combiner functions, Combiner Functions–Specifying a combiner function data flow, Data Flow–Data Flow
running distributed jobs, Running a Distributed MapReduce Job Scan class, Java scheduling in YARN about, Scheduling in YARN
Capacity Scheduler, Capacity Scheduler Configuration–Queue placement delay scheduling, Delay Scheduling
Dominant Resource Fairness, Dominant Resource Fairness
Fair Scheduler, Fair Scheduler Configuration–Preemption FIFO Scheduler, Scheduler Options jobs, Job scheduler
scheduling tasks in Spark, Task Scheduling
schema-on-read, Relational Database Management Systems, Schema on Read Versus Schema on Write
schema-on-write, Schema on Read Versus Schema on Write
schemas
Avro, Avro Data Types and Schemas–Avro Data Types and Schemas, Avro, Protocol Buffers, and Thrift
HBase online query application, Schema Design
MySQL, A Sample Import
Parquet, Writing and Reading Parquet Files
Pig Latin, Schemas–Schema merging, Using a schema ScriptBasedMapping class, Rack awareness scripts
MapReduce, MapReduce Scripts
Pig, Running Pig Programs
Python, MapReduce Scripts
ZooKeeper, Resilience and Performance search platforms, Beyond Batch secondary namenodes about, Namenodes and Datanodes
checkpointing process, The filesystem image and edit log directory structure, Secondary namenode directory structure starting, Starting and Stopping the Daemons
secondary sort, Secondary Sort–Streaming SecondarySort class, Crunch Libraries security about, Security
additional enhancements, Other Security Enhancements–Other Security Enhancements delegation tokens, Delegation Tokens
Kerberos and, Kerberos and Hadoop–An example security.datanode.protocol.acl property, Other Security Enhancements seek time, Relational Database Management Systems
Seekable interface, FSDataInputStream
SELECT statement (Hive)
grouping rows, An Example
index support, Updates, Transactions, and Indexes partitioned data and, Inserts subqueries and, Subqueries views and, Views
SELECT TRANSFORM statement (Hive), User-Defined Functions selectors, replicating and multiplexing, Replicating and Multiplexing Selectors semi joins, Semi joins
semi-structured data, Relational Database Management Systems semicolons, Structure
SequenceFile class about, SequenceFile
compressing streams, Compressing and decompressing streams with CompressionCodec
converting tar files, SequenceFile
displaying with command-line interface, Displaying a SequenceFile with the commandline interface
exports and, Exports and SequenceFiles
format overview, The SequenceFile format–The SequenceFile format
NullWritable class and, NullWritable
ObjectWritable class and, ObjectWritable and GenericWritable reading, Reading a SequenceFile–Reading a SequenceFile sorting and merging, Sorting and merging SequenceFiles Sqoop support, Text and Binary File Formats writing, Writing a SequenceFile–Writing a SequenceFile
SequenceFileAsBinaryInputFormat class, SequenceFileAsBinaryInputFormat
SequenceFileAsBinaryOutputFormat class, SequenceFileAsBinaryOutputFormat
SequenceFileAsTextInputFormat class, SequenceFileAsTextInputFormat
SequenceFileInputFormat class, SequenceFileInputFormat
SequenceFileOutputFormat class, Using Compression in MapReduce, Processing a whole file as a record, SequenceFileOutputFormat sequential znodes, Sequence numbers
SerDe (Serializer-Deserializer), Storage Formats–Using a custom SerDe: RegexSerDe SERDE keyword (Hive), Using a custom SerDe: RegexSerDe
Serializable interface (Java), Serialization of functions serialization about, Serialization–Serialization
Avro support, In-Memory Serialization and Deserialization–The Specific API DefaultStringifier class, Using the Job Configuration of functions, Serialization of functions IDL support, Serialization IDL
implementing custom Writable, Implementing a Custom Writable–Custom comparators pluggable frameworks, Serialization Frameworks–Serialization IDL
RDD, Serialization
Sqoop support, Additional Serialization Systems tuning checklist, Tuning a Job
Writable class hierarchy, Writable Classes–Writable collections
Writable interface, Serialization–WritableComparable and comparators
Serialization interface, Serialization Frameworks Serializer interface, Serialization Frameworks serializer property, File Formats
Serializer-Deserializer (SerDe), Storage Formats–Using a custom SerDe: RegexSerDe service requests, Kerberos and Hadoop
Set class, Crunch Libraries
SET command (Hive), Configuring Hive set command (Pig), Statements setACL operation (ZooKeeper), Operations setData operation (ZooKeeper), Operations
SetFile class, MapFile variants
SETI@home project, Volunteer Computing sh command, Statements Shard class, Crunch Libraries
shared-nothing architecture, Grid Computing
ShareThis sharing network, Hadoop and Cascading at ShareThis–Hadoop and Cascading at ShareThis
short-circuit local reads, Short-circuit local reads
ShortWritable class, Writable wrappers for Java primitives
SHOW FUNCTIONS statement (Hive), Operators and Functions
SHOW LOCKS statement (Hive), Updates, Transactions, and Indexes
SHOW PARTITIONS statement (Hive), Partitions SHOW TABLES statement (Hive), Views shuffle process about, Shuffle and Sort
configuration tuning, Configuration Tuning–Configuration Tuning map side, The Map Side–The Map Side reduce side, The Reduce Side–The Reduce Side SHUFFLED_MAPS counter, Task counters
side data distribution about, Side Data Distribution
distributed cache, Distributed Cache–The distributed cache API job configuration, Using the Job Configuration Sierra, Stuart, SequenceFile
single point of failure (SPOF), HDFS High Availability single sign-ons, Kerberos and Hadoop sink groups (Flume), Sink Groups–Sink Groups sinkgroups property, Sink Groups
SIZE function (Pig Latin), Validation and nulls, Functions
slaves file, Starting and Stopping the Daemons, Hadoop Configuration, Commissioning new nodes
Snappy compression, Compression–Codecs, Native libraries
SnappyCodec class, Codecs
SORT BY clause (Hive), Sorting and Aggregating
Sort class, Crunch, Crunch Libraries
SortedMapWritable class, Writable collections sorting data
about, Sorting
Avro and, Sort Order, Sorting Using Avro MapReduce–Sorting Using Avro MapReduce controlling sort order, Partial Sort
Hive tables, Sorting and Aggregating
MapReduce and, Sorting–Streaming, Sorting Using Avro MapReduce–Sorting Using
Avro MapReduce
partial sort, Partial Sort–Partial Sort Pig operators and, Sorting Data preparation overview, Preparation secondary sort, Secondary Sort–Streaming
shuffle process and, Shuffle and Sort–Configuration Tuning total sort, Total Sort–Total Sort
Source interface, Reading from a source
SourceTarget interface, Combined sources and targets
Spark about, Spark
additonal information, Further Reading
anatomy of job runs, Anatomy of a Spark Job Run–Task Execution cluster managers and, Executors and Cluster Managers example of, An Example–A Python Example executors and, Executors and Cluster Managers Hive and, Execution engines installing, Installing Spark
MapReduce and, Transformations and Actions RDDs and, Resilient Distributed Datasets–Functions resource requests, Resource Requests shared variables, Shared Variables–Accumulators sorting data, Total Sort
YARN and, Spark on YARN–YARN cluster mode spark-shell command, An Example
spark-submit command, A Scala Standalone Application, YARN cluster mode spark.kryo.registrator property, Data
SparkConf class, A Scala Standalone Application
SparkContext class, An Example, YARN client mode–YARN cluster mode SpecificDatumReader class, The Specific API
speculative execution of tasks, Speculative Execution–Speculative Execution
SPILLED_RECORDS counter, Task counters
SPLIT statement (Pig Latin), Statements, Combining and Splitting Data splits (input data) (see input splits)
SPLIT_RAW_BYTES counter, Task counters
SPOF (single point of failure), HDFS High Availability
Sqoop about, Sqoop
additional information, Further Reading Avro support, Text and Binary File Formats connectors and, Sqoop Connectors
escape sequences supported, Performing an Export
export process, Performing an Export–Exports and SequenceFiles file formats, Text and Binary File Formats generated code, Generated Code getting, Getting Sqoop–Getting Sqoop
import process, Imports: A Deeper Look–Direct-Mode Imports importing large objects, Importing Large Objects–Importing Large Objects
MapReduce support, A Sample Import, Imports: A Deeper Look, Exports: A Deeper Look
Parquet support, Text and Binary File Formats
sample import, A Sample Import–Text and Binary File Formats SequenceFile class and, Text and Binary File Formats serialization and, Additional Serialization Systems tool support, Getting Sqoop, Generated Code
working with imported data, Working with Imported Data–Imported Data and Hive
srst command (ZooKeeper), Installing and Running ZooKeeper srvr command (ZooKeeper), Installing and Running ZooKeeper SSH, configuring, Configuring SSH, SSH settings, Configuring SSH stack traces, Getting stack traces
Stack, Michael, HBasics–Further Reading
standalone mode (Hadoop), Standalone Mode
stat command (ZooKeeper), Installing and Running ZooKeeper statements (Pig Latin) about, Statements–Statements control flow, Statements
expressions and, Expressions–Expressions
states (ZooKeeper), States–States, State exceptions status updates for tasks, Progress and Status Updates storage handlers, Storage handlers store functions (Pig Latin), Functions
STORE statement (Pig Latin), Statements, Statements, Sorting Data
STORED AS clause (Hive), Binary storage formats: Sequence files, Avro datafiles, Parquet files, RCFiles, and ORCFiles
STORED BY clause (Hive), Storage handlers
STREAM statement (Pig Latin), Statements, STREAM
stream.map.input.field.separator property, Keys and values in Streaming stream.map.input.ignoreKey property, The default Streaming job stream.map.output.field.separator property, Keys and values in Streaming stream.non.zero.exit.is.failure property, Task Failure
stream.num.map.output.key.fields property, Keys and values in Streaming stream.num.reduce.output.key.fields property, Keys and values in Streaming stream.recordreader.class property, XML
stream.reduce.input.field.separator property, Keys and values in Streaming stream.reduce.output.field.separator property, Keys and values in Streaming
Streaming programs about, Beyond Batch
default job, The default Streaming job–Keys and values in Streaming secondary sort, Streaming–Streaming task execution, Streaming
user-defined counters, User-Defined Streaming Counters
StreamXmlRecordReader class, XML
StrictHostKeyChecking SSH setting, SSH settings
String class (Java), Text–Resorting to String, Avro Data Types and Schemas StringTokenizer class (Java), MapReduce Library Classes StringUtils class, The Writable Interface, Dynamic invokers structured data, Relational Database Management Systems subqueries, Subqueries
SUM function (Pig Latin), Functions
SWebHdfsFileSystem class, Hadoop Filesystems
SwiftNativeFileSystem class, Hadoop Filesystems SWIM repository, Other benchmarks sync operation (ZooKeeper), Operations syncLimit property, Configuration syslog file (Java), Hadoop Logs system administration commissioning nodes, Commissioning and Decommissioning Nodes–Commissioning new nodes
decommissioning nodes, Decommissioning old nodes–Decommissioning old nodes HDFS support, Persistent Data Structures–Balancer monitoring, Monitoring–Metrics and JMX
routine procedures, Metadata backups–Filesystem balancer upgrading clusters, Upgrades–Finalize the upgrade (optional) System class (Java), GenericOptionsParser, Tool, and ToolRunner system logfiles, Hadoop Logs, System logfiles
T
TableInputFormat class, Database Input (and Output), MapReduce
TableMapper class, MapReduce
TableMapReduceUtil class, MapReduce
TableOutputFormat class, Database Input (and Output), MapReduce tables (HBase)
about, Whirlwind Tour of the Data Model–Whirlwind Tour of the Data Model creating, Test Drive inserting data into, Test Drive locking, Locking regions, Regions removing, Test Drive wide tables, Schema Design
tables (Hive) about, Tables altering, Altering Tables
buckets and, Partitions and Buckets, Buckets–Buckets dropping, Dropping Tables
external tables, Managed Tables and External Tables–Managed Tables and External Tables
importing data, Importing Data–CREATE TABLE…AS SELECT
managed tables, Managed Tables and External Tables–Managed Tables and External Tables
partitions and, Partitions and Buckets–Partitions storage formats, Storage Formats–Storage handlers views, Views
TABLESAMPLE clause (Hive), Buckets
TableSource interface, Reading from a source Target interface, Writing to a target
task attempt IDs, Launching a Job, The Task Execution Environment task attempts page (MapReduce), The tasks and task attempts pages task counters, Built-in Counters–Task counters
task IDs, Launching a Job, The Task Execution Environment task logs (MapReduce), Hadoop Logs
TaskAttemptContext interface, Progress and Status Updates tasks
executing, Task Execution, Task Execution–Task side-effect files, Task Execution failure considerations, Task Failure profiling, Profiling Tasks–The HPROF profiler progress and status updates, Progress and Status Updates scheduling in Spark, Task Scheduling
Spark support, Spark Applications, Jobs, Stages, and Tasks speculative execution, Speculative Execution–Speculative Execution streaming, Streaming
task assignments, Task Assignment
tasks page (MapReduce), The tasks and task attempts pages tasktrackers, YARN Compared to MapReduce 1 TEMPORARY keyword (Hive), Writing a UDF teragen program, Benchmarking MapReduce with TeraSort
TeraSort program, Benchmarking MapReduce with TeraSort TestDFSIO benchmark, Other benchmarks testing
HBase installation, Test Drive–Test Drive Hive considerations, The Hive Shell
job drivers, Testing the Driver–Testing the Driver MapReduce test runs, A test run–A test run in miniclusters, Testing the Driver
running jobs locally on test data, Running Locally on Test Data–Testing the Driver running jobs on clusters, Running on a Cluster–Remote Debugging writing unit tests with MRUnit, Writing a Unit Test with MRUnit–Reducer
Text class, Text–Resorting to String, Implementing a Custom Writable–Implementing a
RawComparator for speed, MapReduce Types text formats
controlling maximum line length, Controlling the maximum line length
KeyValueTextInputFormat class, KeyValueTextInputFormat
NLineInputFormat class, NLineInputFormat
NullOutputFormat class, Text Output
TextInputFormat class, TextInputFormat
TextOutputFormat class, Text Output
XML documents and, XML
TextInputFormat class about, TextInputFormat
MapReduce types and, Running a Job in a Local Job Runner, MapReduce Types Sqoop imports and, Working with Imported Data
TextLoader function (Pig Latin), Functions
TextOutputFormat class, Implementing a Custom Writable, Text Output, An Example TGT (Ticket-Granting Ticket), Kerberos and Hadoop thread dumps, Getting stack traces
Thrift
HBase and, REST and Thrift
Hive and, Hive clients
Parquet and, Avro, Protocol Buffers, and Thrift–Projection and read schemas ThriftParquetWriter class, Avro, Protocol Buffers, and Thrift tick time (ZooKeeper), Time
Ticket-Granting Ticket (TGT), Kerberos and Hadoop timeline servers, YARN Compared to MapReduce 1 TOBAG function (Pig Latin), Types, Functions tojson command, Avro Tools
TokenCounterMapper class, MapReduce Library Classes
TOKENSIZE function (Pig Latin), Functions
ToLowerFn function, Materialization
TOMAP function (Pig Latin), Types, Functions
Tool interface, GenericOptionsParser, Tool, and ToolRunner–GenericOptionsParser, Tool, and ToolRunner
ToolRunner class, GenericOptionsParser, Tool, and ToolRunner–GenericOptionsParser, Tool, and ToolRunner
TOP function (Pig Latin), Functions
TotalOrderPartitioner class, Total Sort
TOTAL_LAUNCHED_MAPS counter, Job counters
TOTAL_LAUNCHED_REDUCES counter, Job counters
TOTAL_LAUNCHED_UBERTASKS counter, Job counters
TOTUPLE function (Pig Latin), Types, Functions TPCx-HS benchmark, Other benchmarks
transfer rate, Relational Database Management Systems TRANSFORM clause (Hive), MapReduce Scripts
transformations, RDD, Transformations and Actions–Aggregation transformations Trash class, Trash trash facility, Trash
TRUNCATE TABLE statement (Hive), Dropping Tables tuning jobs, Tuning a Job–The HPROF profiler
TwoDArrayWritable class, Writable collections
U
uber tasks, Job Initialization
UDAF class, Writing a UDAF
UDAFEvaluator interface, Writing a UDAF
UDAFs (user-defined aggregate functions), User-Defined Functions, Writing a UDAF–A more complex UDAF
UDF class, Writing a UDF
UDFs (user-defined functions)
Hive and, User-Defined Functions–A more complex UDAF
Pig and, Pig, Other libraries, User-Defined Functions–Using a schema
UDTFs (user-defined table-generating functions), User-Defined Functions
Unicode characters, Unicode–Unicode
UNION statement (Pig Latin), Statements, Combining and Splitting Data
unit tests with MRUnit, Setting Up the Development Environment, Writing a Unit Test with MRUnit–Reducer
Unix user accounts, Creating Unix User Accounts
unmanaged application masters, Anatomy of a YARN Application Run unstructured data, Relational Database Management Systems
UPDATE statement (Hive), Updates, Transactions, and Indexes upgrading clusters, Upgrades–Finalize the upgrade (optional) URL class (Java), Reading Data from a Hadoop URL user accounts, Unix, Creating Unix User Accounts user identity, Managing Configuration
user-defined aggregate functions (UDAFs), User-Defined Functions, Writing a UDAF–A more complex UDAF
user-defined functions (see UDFs)
user-defined table-generating functions (UDTFs), User-Defined Functions USING JAR clause (Hive), Writing a UDF
V
VCORES_MILLIS_MAPS counter, Job counters
VCORES_MILLIS_REDUCES counter, Job counters VERSION file, Namenode directory structure versions (Hive), Installing Hive
ViewFileSystem class, HDFS Federation, Hadoop Filesystems views (virtual tables), Views
VIntWritable class, Writable wrappers for Java primitives
VIRTUAL_MEMORY_BYTES counter, Task counters, Memory settings in YARN and MapReduce
VLongWritable class, Writable wrappers for Java primitives volunteer computing, Volunteer Computing
W
w (write) permission, Basic Filesystem Operations
Walters, Chad, Backdrop
WAR (Web application archive) files, Packaging a Job watches (ZooKeeper), Watches, Watch triggers Watson, James D., The Structure of DNA
wchc command (ZooKeeper), Installing and Running ZooKeeper wchp command (ZooKeeper), Installing and Running ZooKeeper wchs command (ZooKeeper), Installing and Running ZooKeeper
Web application archive (WAR) files, Packaging a Job
WebHDFS protocol, HTTP
WebHdfsFileSystem class, Hadoop Filesystems webtables (HBase), HBasics
Wensel, Chris K., Cascading
Whitacre, Micah, Composable Data at Cerner whoami command, Managing Configuration
WITH SERDEPROPERTIES clause (Hive), Using a custom SerDe: RegexSerDe work units, Volunteer Computing, Data Flow workflow engines, Apache Oozie workflows (MapReduce)
about, MapReduce Workflows
Apache Oozie system, Apache Oozie–Running an Oozie workflow job decomposing problems into jobs, Decomposing a Problem into MapReduce Jobs–Decomposing a Problem into MapReduce Jobs JobControl class, JobControl
Writable interface about, Serialization–WritableComparable and comparators class hierarchy, Writable Classes–Writable collections Crunch and, Types
implementing custom, Implementing a Custom Writable–Custom comparators
WritableComparable interface, WritableComparable and comparators, Partial Sort
WritableComparator class, WritableComparable and comparators
WritableSerialization class, Serialization Frameworks WritableUtils class, Custom comparators write (w) permission, Basic Filesystem Operations
WRITE permission (ACL), ACLs
WriteSupport class, Writing and Reading Parquet Files WRITE_OPS counter, Task counters writing data
Crunch support, Writing to a target
using FileSystem API, Writing Data–FSDataOutputStream
HDFS data flow, Anatomy of a File Write–Anatomy of a File Write
Parquet and, Writing and Reading Parquet Files–Projection and read schemas
SequenceFile class, Writing a SequenceFile–Writing a SequenceFile
X
x (execute) permission, Basic Filesystem Operations
XML documents, XML
Y
Yahoo!, A Brief History of Apache Hadoop
YARN (Yet Another Resource Negotiator) about, Beyond Batch, YARN, Further Reading
anatomy of application run, Anatomy of a YARN Application Run–Building YARN Applications
application lifespan, Application Lifespan application master failure, Application Master Failure building applications, Building YARN Applications cluster setup and installation, Creating Unix User Accounts cluster sizing, Master node scenarios
daemon properties, YARN–CPU settings in YARN and MapReduce distributed shell, Building YARN Applications log aggregation, Hadoop Logs
MapReduce comparison, YARN Compared to MapReduce 1–YARN Compared to
MapReduce 1
scaling out data, Scaling Out
scheduling in, Scheduling in YARN–Dominant Resource Fairness, Job scheduler Spark and, Spark on YARN–YARN cluster mode
starting and stopping daemons, Starting and Stopping the Daemons
YARN client mode (Spark), Spark on YARN
YARN cluster mode (Spark), YARN cluster mode–YARN cluster mode yarn-env.sh file, Hadoop Configuration
yarn-site.xml file, Hadoop Configuration, Important Hadoop Daemon Properties yarn.app.mapreduce.am.job.recovery.enable property, Application Master Failure yarn.app.mapreduce.am.job.speculator.class property, Speculative Execution yarn.app.mapreduce.am.job.task.estimator.class property, Speculative Execution yarn.log-aggregation-enable property, Hadoop Logs
yarn.nodemanager.address property, Hadoop Daemon Addresses and Ports yarn.nodemanager.aux-services property, YARN, Configuration yarn.nodemanager.bind-host property, Hadoop Daemon Addresses and Ports
yarn.nodemanager.container-executor.class property, Task Failure, CPU settings in YARN and MapReduce, Other Security Enhancements
yarn.nodemanager.delete.debug-delay-sec property, Remote Debugging yarn.nodemanager.hostname property, Hadoop Daemon Addresses and Ports yarn.nodemanager.linux-container-executor property, CPU settings in YARN and MapReduce
yarn.nodemanager.local-dirs property, YARN
yarn.nodemanager.localizer.address property, Hadoop Daemon Addresses and Ports yarn.nodemanager.log.retain-second property, Hadoop Logs
yarn.nodemanager.resource.cpu-vcores property, YARN, CPU settings in YARN and MapReduce
yarn.nodemanager.resource.memory-mb property, GenericOptionsParser, Tool, and ToolRunner, YARN
yarn.nodemanager.vmem-pmem-ratio property, YARN, Memory settings in YARN and MapReduce
yarn.nodemanager.webapp.address property, Hadoop Daemon Addresses and Ports yarn.resourcemanager.address property about, YARN, Hadoop Daemon Addresses and Ports
Hive and, Configuring Hive
Pig and, MapReduce mode yarn.resourcemanager.admin.address property, Hadoop Daemon Addresses and Ports yarn.resourcemanager.am.max-attempts property, Application Master Failure, Resource Manager Failure
yarn.resourcemanager.bind-host property, Hadoop Daemon Addresses and Ports yarn.resourcemanager.hostname property, YARN, Hadoop Daemon Addresses and Ports, Configuration
yarn.resourcemanager.max-completed-applications property, The resource manager page yarn.resourcemanager.nm.liveness-monitor.expiry-interval-ms property, Node Manager Failure
yarn.resourcemanager.nodes.exclude-path property, Cluster membership, Decommissioning old nodes
yarn.resourcemanager.nodes.include-path property, Cluster membership, Commissioning new nodes
yarn.resourcemanager.resource-tracker.address property, Hadoop Daemon Addresses and Ports
yarn.resourcemanager.scheduler.address property, Hadoop Daemon Addresses and Ports yarn.resourcemanager.scheduler.class property, Enabling the Fair Scheduler yarn.resourcemanager.webapp.address property, Hadoop Daemon Addresses and Ports yarn.scheduler.capacity.node-locality-delay property, Delay Scheduling yarn.scheduler.fair.allocation.file property, Queue configuration yarn.scheduler.fair.allow-undeclared-pools property, Queue placement yarn.scheduler.fair.locality.threshold.node property, Delay Scheduling yarn.scheduler.fair.locality.threshold.rack property, Delay Scheduling yarn.scheduler.fair.preemption property, Preemption
yarn.scheduler.fair.user-as-default-queue property, Queue placement
yarn.scheduler.maximum-allocation-mb property, Memory settings in YARN and MapReduce
yarn.scheduler.minimum-allocation-mb property, Memory settings in YARN and MapReduce
yarn.web-proxy.address property, Hadoop Daemon Addresses and Ports
YARN_LOG_DIR environment variable, Hadoop Logs
YARN_RESOURCEMANAGER_HEAPSIZE environment variable, Memory heap size
Z
Zab protocol, Implementation zettabytes, Data!
znodes about, Group Membership in ZooKeeper ACLs and, ACLs
creating, Creating the Group–Creating the Group deleting, Deleting a Group ephemeral, Ephemeral znodes joining groups, Joining a Group
listing , Listing Members in a Group–ZooKeeper command-line tools operations supported, Operations persistent, Ephemeral znodes
properties supported, Data Model–Watches sequential, Sequence numbers
ZOOCFGDIR environment variable, Installing and Running ZooKeeper
ZooKeeper about, ZooKeeper
additional information, Further Reading building applications configuration service, A Configuration Service–A Configuration Service, A Lock Service–Implementation
distributed data structures and protocols, More Distributed Data Structures and Protocols
resilient, The Resilient ZooKeeper Application–A reliable configuration service
consistency and, Consistency–Consistency data model, Data Model
example of, An Example–Deleting a Group failover controllers and, Failover and fencing HBase and, Implementation
high availability and, HDFS High Availability implementing, Implementation
installing and running, Installing and Running ZooKeeper–Installing and Running ZooKeeper
operations in, Operations–ACLs
production considerations, ZooKeeper in Production–Configuration sessions and, Sessions–Time
states and, States–States, State exceptions zxid, Consistency
Colophon
The animal on the cover of Hadoop: The Definitive Guide is an African elephant. These members of the genus Loxodonta are the largest land animals on Earth (slightly larger than their cousin, the Asian elephant) and can be identified by their ears, which have been said to look somewhat like the continent of Asia. Males stand 12 feet tall at the shoulder and weigh 12,000 pounds, but they can get as big as 15,000 pounds, whereas females stand 10 feet tall and weigh 8,000–11,000 pounds. Even young elephants are very large: at birth, they already weigh approximately 200 pounds and stand about 3 feet tall.
African elephants live throughout sub-Saharan Africa. Most of the continent’s elephants live on savannas and in dry woodlands. In some regions, they can be found in desert areas; in others, they are found in mountains.
The species plays an important role in the forest and savanna ecosystems in which they live. Many plant species are dependent on passing through an elephant’s digestive tract before they can germinate; it is estimated that at least a third of tree species in west African forests rely on elephants in this way. Elephants grazing on vegetation also affect the structure of habitats and influence bush fire patterns. For example, under natural conditions, elephants make gaps through the rainforest, enabling the sunlight to enter, which allows the growth of various plant species. This, in turn, facilitates more abundance and more diversity of smaller animals. As a result of the influence elephants have over many plants and animals, they are often referred to as a keystone species because they are vital to the long-term survival of the ecosystems in which they live.
Many of the animals on O’Reilly covers are endangered; all of them are important to the world. To learn more about how you can help, go to animals.oreilly.com.
The cover image is from the Dover Pictorial Archive. The cover fonts are URW Typewriter and Guardian Sans. The text font is Adobe Minion Pro; the heading font is Adobe Myriad Condensed; and the code font is Dalton Maag’s Ubuntu Mono.
Hadoop: The Definitive Guide
Tom White
Editor
Mike Loukides
Editor
Meghan Blanchette
Editor
Matthew Hacker
Editor
Jasmine Kwityn
Revision History
| 2015-03-19 | First release |
|---|---|
Copyright © 2015 Tom White
O’Reilly books may be purchased for educational, business, or sales promotional use. Online editions are also available for most titles (http://safaribooksonline.com). For more information, contact our corporate/institutional sales department: 800-998-9938 or corporate@oreilly.com.
The O’Reilly logo is a registered trademark of O’Reilly Media, Inc. Hadoop: The Definitive Guide, the cover image of an African elephant, and related trade dress are trademarks of O’Reilly Media, Inc.
Many of the designations used by manufacturers and sellers to distinguish their products are claimed as trademarks. Where those designations appear in this book, and O’Reilly Media, Inc. was aware of a trademark claim, the designations have been printed in caps or initial caps.
While the publisher and the author have used good faith efforts to ensure that the information and instructions contained in this work are accurate, the publisher and the author disclaim all responsibility for errors or omissions, including without limitation responsibility for damages resulting from the use of or reliance on this work. Use of the information and instructions contained in this work is at your own risk. If any code samples or other technology this work contains or describes is subject to open source licenses or the intellectual property rights of others, it is your responsibility to ensure that your use thereof complies with such licenses and/or rights.
O’Reilly Media, Inc.
1005 Gravenstein Highway North
Sebastopol, CA 95472
2015-03-19T19:44:25Z
Hadoop: The Definitive Guide
Table of Contents
Dedication
Foreword
Preface
Administrative Notes What’s New in the Fourth Edition?
What’s New in the Third Edition?
What’s New in the Second Edition?
Conventions Used in This Book
Using Code Examples
Safari® Books Online
How to Contact Us
Acknowledgments
I. Hadoop Fundamentals 1. Meet Hadoop
Data!
Data Storage and Analysis
Querying All Your Data
Beyond Batch
Comparison with Other Systems
Relational Database Management Systems
Grid Computing
Volunteer Computing
A Brief History of Apache Hadoop What’s in This Book?
2. MapReduce
A Weather Dataset
Data Format
Analyzing the Data with Unix Tools
Analyzing the Data with Hadoop
Map and Reduce
Java MapReduce
A test run
Scaling Out
Data Flow
Combiner Functions
Specifying a combiner function
Running a Distributed MapReduce Job
Hadoop Streaming
Ruby
Python
3. The Hadoop Distributed Filesystem
The Design of HDFS
HDFS Concepts
Blocks
Namenodes and Datanodes
Block Caching
HDFS Federation
HDFS High Availability
Failover and fencing
The Command-Line Interface
Basic Filesystem Operations
Hadoop Filesystems
Interfaces
HTTP
C
NFS
FUSE
The Java Interface
Reading Data from a Hadoop URL
Reading Data Using the FileSystem API FSDataInputStream
Writing Data
FSDataOutputStream
Directories
Querying the Filesystem
File metadata: FileStatus
Listing files
File patterns
PathFilter
Deleting Data
Data Flow
Anatomy of a File Read
Anatomy of a File Write
Coherency Model
Consequences for application design
Parallel Copying with distcp
Keeping an HDFS Cluster Balanced
4. YARN
Anatomy of a YARN Application Run
Resource Requests
Application Lifespan
Building YARN Applications
YARN Compared to MapReduce 1
Scheduling in YARN
Scheduler Options
Capacity Scheduler Configuration
Queue placement
Fair Scheduler Configuration
Enabling the Fair Scheduler
Queue configuration
Queue placement
Preemption
Delay Scheduling
Dominant Resource Fairness
Further Reading
5. Hadoop I/O Data Integrity
Data Integrity in HDFS
LocalFileSystem
ChecksumFileSystem
Compression
Codecs
Compressing and decompressing streams with CompressionCodec
Inferring CompressionCodecs using CompressionCodecFactory
Native libraries
CodecPool
Compression and Input Splits
Using Compression in MapReduce
Compressing map output
Serialization
The Writable Interface
WritableComparable and comparators
Writable Classes
Writable wrappers for Java primitives
Text
Indexing
Unicode
Iteration
Mutability
Resorting to String
BytesWritable
NullWritable
ObjectWritable and GenericWritable
Writable collections
Implementing a Custom Writable
Implementing a RawComparator for speed
Custom comparators
Serialization Frameworks Serialization IDL
File-Based Data Structures
SequenceFile
Writing a SequenceFile
Reading a SequenceFile
Displaying a SequenceFile with the command-line interface
Sorting and merging SequenceFiles
The SequenceFile format
MapFile
MapFile variants
Other File Formats and Column-Oriented Formats
II. MapReduce
6. Developing a MapReduce Application
The Configuration API
Combining Resources
Variable Expansion
Setting Up the Development Environment
Managing Configuration
GenericOptionsParser, Tool, and ToolRunner
Writing a Unit Test with MRUnit
Mapper
Reducer
Running Locally on Test Data
Running a Job in a Local Job Runner
Testing the Driver
Running on a Cluster
Packaging a Job
The client classpath
The task classpath
Packaging dependencies
Task classpath precedence
Launching a Job
The MapReduce Web UI
The resource manager page
The MapReduce job page
Retrieving the Results
Debugging a Job
The tasks and task attempts pages
Handling malformed data
Hadoop Logs
Remote Debugging
Tuning a Job
Profiling Tasks
The HPROF profiler
MapReduce Workflows
Decomposing a Problem into MapReduce Jobs
JobControl
Apache Oozie
Defining an Oozie workflow
Packaging and deploying an Oozie workflow application
Running an Oozie workflow job 7. How MapReduce Works
Anatomy of a MapReduce Job Run
Job Submission
Job Initialization
Task Assignment
Task Execution
Streaming
Progress and Status Updates
Job Completion
Failures
Task Failure
Application Master Failure
Node Manager Failure
Resource Manager Failure
Shuffle and Sort
The Map Side
The Reduce Side
Configuration Tuning
Task Execution
The Task Execution Environment
Streaming environment variables
Speculative Execution
Output Committers
Task side-effect files
8. MapReduce Types and Formats
MapReduce Types
The Default MapReduce Job
The default Streaming job
Keys and values in Streaming
Input Formats
Input Splits and Records
FileInputFormat
FileInputFormat input paths
FileInputFormat input splits
Small files and CombineFileInputFormat
Preventing splitting
File information in the mapper
Processing a whole file as a record
Text Input
TextInputFormat
Controlling the maximum line length
KeyValueTextInputFormat
NLineInputFormat
XML
Binary Input
SequenceFileInputFormat
SequenceFileAsTextInputFormat
SequenceFileAsBinaryInputFormat
FixedLengthInputFormat
Multiple Inputs
Database Input (and Output)
Output Formats
Text Output
Binary Output
SequenceFileOutputFormat
SequenceFileAsBinaryOutputFormat MapFileOutputFormat
Multiple Outputs
An example: Partitioning data
MultipleOutputs
Lazy Output
Database Output
9. MapReduce Features
Counters
Built-in Counters
Task counters
Job counters
User-Defined Java Counters
Dynamic counters
Retrieving counters
User-Defined Streaming Counters
Sorting
Preparation
Partial Sort
Total Sort
Secondary Sort
Java code
Streaming
Joins
Map-Side Joins
Reduce-Side Joins
Side Data Distribution
Using the Job Configuration
Distributed Cache
Usage
How it works
The distributed cache API
MapReduce Library Classes
III. Hadoop Operations
10. Setting Up a Hadoop Cluster
Cluster Specification
Cluster Sizing
Master node scenarios
Network Topology
Rack awareness
Cluster Setup and Installation
Installing Java
Creating Unix User Accounts
Installing Hadoop
Configuring SSH
Configuring Hadoop
Formatting the HDFS Filesystem
Starting and Stopping the Daemons
Creating User Directories
Hadoop Configuration
Configuration Management
Environment Settings
Java
Memory heap size
System logfiles
SSH settings
Important Hadoop Daemon Properties
HDFS
YARN
Memory settings in YARN and MapReduce
CPU settings in YARN and MapReduce
Hadoop Daemon Addresses and Ports
Other Hadoop Properties
Cluster membership
Buffer size
HDFS block size
Reserved storage space
Trash
Job scheduler
Reduce slow start
Short-circuit local reads
Security
Kerberos and Hadoop
An example
Delegation Tokens
Other Security Enhancements
Benchmarking a Hadoop Cluster
Hadoop Benchmarks
Benchmarking MapReduce with TeraSort
Other benchmarks
User Jobs
11. Administering Hadoop
HDFS
Persistent Data Structures
Namenode directory structure
The filesystem image and edit log
Secondary namenode directory structure
Datanode directory structure
Safe Mode
Entering and leaving safe mode
Audit Logging
Tools dfsadmin
Filesystem check (fsck)
Finding the blocks for a file
Datanode block scanner
Balancer
Monitoring
Logging
Setting log levels
Getting stack traces
Metrics and JMX
Maintenance
Routine Administration Procedures
Metadata backups
Data backups
Filesystem check (fsck)
Filesystem balancer
Commissioning and Decommissioning Nodes
Commissioning new nodes
Decommissioning old nodes
Upgrades
HDFS data and metadata upgrades
Start the upgrade
Wait until the upgrade is complete
Check the upgrade
Roll back the upgrade (optional)
Finalize the upgrade (optional)
IV. Related Projects
12. Avro
Avro Data Types and Schemas
In-Memory Serialization and Deserialization The Specific API
Avro Datafiles
Interoperability
Python API
Avro Tools
Schema Resolution
Sort Order
Avro MapReduce
Sorting Using Avro MapReduce
Avro in Other Languages
13. ParquetData Model
Nested Encoding
Parquet File Format
Parquet Configuration
Writing and Reading Parquet Files
Avro, Protocol Buffers, and Thrift
Projection and read schemas
Parquet MapReduce
14. Flume
Installing Flume
An Example
Transactions and Reliability
Batching
The HDFS Sink
Partitioning and Interceptors
File Formats
Fan Out
Delivery Guarantees
Replicating and Multiplexing Selectors
Distribution: Agent Tiers Delivery Guarantees
Sink Groups
Integrating Flume with Applications
Component Catalog
Further Reading
15. Sqoop
Getting Sqoop
Sqoop Connectors
A Sample Import
Text and Binary File Formats
Generated Code
Additional Serialization Systems
Imports: A Deeper Look
Controlling the Import
Imports and Consistency
Incremental Imports
Direct-Mode Imports
Working with Imported Data Imported Data and Hive
Importing Large Objects Performing an Export
Exports: A Deeper Look
Exports and Transactionality Exports and SequenceFiles
Further Reading
16. Pig
Installing and Running Pig
Execution Types
Local mode
MapReduce mode
Running Pig Programs
Grunt
Pig Latin Editors
An Example
Generating Examples
Comparison with Databases
Pig Latin
Structure
Statements
Expressions
Types
Schemas
Using Hive tables with HCatalog
Validation and nulls Schema merging
Functions
Other libraries
Macros
User-Defined Functions
A Filter UDF
Leveraging types
An Eval UDF
Dynamic invokers
A Load UDF
Using a schema
Data Processing Operators
Loading and Storing Data
Filtering Data
FOREACH…GENERATE STREAM
Grouping and Joining Data
JOIN
COGROUP
CROSS
GROUP
Sorting Data
Combining and Splitting Data
Pig in Practice
Parallelism
Anonymous Relations
Parameter Substitution
Dynamic parameters
Parameter substitution processing
Further Reading
17. Hive
Installing Hive
The Hive Shell
An Example
Running Hive
Configuring Hive
Execution engines
Logging
Hive Services Hive clients
The Metastore
Comparison with Traditional Databases
Schema on Read Versus Schema on Write
Updates, Transactions, and Indexes
SQL-on-Hadoop Alternatives
HiveQL
Data Types
Primitive types
Complex types
Operators and Functions
Conversions
Tables
Managed Tables and External Tables
Partitions and Buckets
Partitions
Buckets
Storage Formats
The default storage format: Delimited text
Binary storage formats: Sequence files, Avro datafiles, Parquet files, RCFiles, and ORCFiles
Using a custom SerDe: RegexSerDe
Storage handlers
Importing Data
Inserts
Multitable insert
CREATE TABLE…AS SELECT
Altering Tables
Dropping Tables
Querying Data
Sorting and Aggregating
MapReduce Scripts
Joins
Inner joins
Outer joins Semi joins
Map joins
Subqueries
Views
User-Defined Functions
Writing a UDF
Writing a UDAF
A more complex UDAF
Further Reading
18. Crunch
An Example
The Core Crunch API
Primitive Operations union() parallelDo() groupByKey() combineValues() Types
Records and tuples
Sources and Targets
Reading from a source
Writing to a target
Existing outputs
Combined sources and targets
Functions
Serialization of functions
Object reuse
Materialization PObject
Pipeline Execution
Running a Pipeline
Asynchronous execution Debugging
Stopping a Pipeline
Inspecting a Crunch Plan Iterative Algorithms
Checkpointing a Pipeline
Crunch Libraries
Further Reading
19. Spark
Installing Spark
An Example
Spark Applications, Jobs, Stages, and Tasks
A Scala Standalone Application
A Java Example
A Python Example
Resilient Distributed Datasets
Creation
Transformations and Actions
Aggregation transformations
Persistence
Persistence levels
Serialization
Data
Functions
Shared Variables
Broadcast Variables
Accumulators
Anatomy of a Spark Job Run
Job Submission
DAG Construction
Task Scheduling
Task Execution
Executors and Cluster Managers
Spark on YARN
YARN client mode
YARN cluster mode
Further Reading
20. HBase
HBasics
Backdrop
Concepts
Whirlwind Tour of the Data Model
Regions
Locking
Implementation
HBase in operation
Installation
Test Drive
Clients
Java
MapReduce
REST and Thrift
Building an Online Query Application
Schema Design
Loading Data
Load distribution
Bulk load
Online Queries
Station queries
Observation queries
HBase Versus RDBMS
Successful Service
HBase
Praxis
HDFS
UI
Metrics
Counters
Further Reading
21. ZooKeeper
Installing and Running ZooKeeper
An Example
Group Membership in ZooKeeper
Creating the Group
Joining a Group
Listing Members in a Group
ZooKeeper command-line tools
Deleting a Group
The ZooKeeper Service
Data Model
Ephemeral znodes
Sequence numbers
Watches
Operations
Multiupdate
APIs
Watch triggers
ACLs
Implementation
Consistency
Sessions Time
States
Building Applications with ZooKeeper
A Configuration Service
The Resilient ZooKeeper Application
InterruptedException
KeeperException
State exceptions
Recoverable exceptions
Unrecoverable exceptions
A reliable configuration service
A Lock Service
The herd effect
Recoverable exceptions
Unrecoverable exceptions
Implementation
More Distributed Data Structures and Protocols
BookKeeper and Hedwig
ZooKeeper in Production
Resilience and Performance
Configuration
Further Reading
V. Case Studies
22. Composable Data at Cerner
From CPUs to Semantic Integration
Enter Apache Crunch
Building a Complete Picture
Integrating Healthcare Data
Composability over Frameworks
Moving Forward
23. Biological Data Science: Saving Lives with Software
The Structure of DNA
The Genetic Code: Turning DNA Letters into Proteins
Thinking of DNA as Source Code
The Human Genome Project and Reference Genomes
Sequencing and Aligning DNA
ADAM, A Scalable Genome Analysis Platform
Literate programming with the Avro interface description language (IDL)
Column-oriented access with Parquet
A simple example: k-mer counting using Spark and ADAM
From Personalized Ads to Personalized Medicine Join In
24. Cascading
Fields, Tuples, and Pipes
Operations
Taps, Schemes, and Flows
Cascading in Practice
Flexibility
Hadoop and Cascading at ShareThis
Summary
A. Installing Apache Hadoop
Prerequisites
Installation
Configuration
Standalone Mode
Pseudodistributed Mode
Configuring SSH
Formatting the HDFS filesystem
Starting and stopping the daemons
Creating a user directory
Fully Distributed Mode
B. Cloudera’s Distribution Including Apache Hadoop
C. Preparing the NCDC Weather Data
D. The Old and New Java MapReduce APIs
Index
Colophon
Copyright

