DESeq2常用于识别差异基因,它主要使用了标准化因子标准化数据,再根据广义线性模型判别组间差异(组间残差是否显著判断)。在获取差异基因结果后,我们可以进行下一步的富集分析,常用方法有基于在线网站DAVID以及脚本处理的两类,本文介绍基于fgsea的方法计算富集分析得分。更多知识分享请到 https://zouhua.top/。
DEseq2
可参考个人博客差异表达分析之Deseq2了解DESeq2如何标准化数据和识别差异基因。下面给出简要代码
library(DESeq2)library(airway)data("airway")ddsSE <- DESeqDataSet(airway, design = ~ cell + dex)ddsSE <- DESeq(ddsSE)res <- results(ddsSE, tidy = TRUE) %>% na.omit() %>% as_tibble()head(res)
# A tibble: 6 x 7row baseMean log2FoldChange lfcSE stat pvalue padj<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>1 ENSG00000000003 709. 0.381 0.101 3.79 0.000152 0.001282 ENSG00000000419 520. -0.207 0.112 -1.84 0.0653 0.1973 ENSG00000000457 237. -0.0379 0.143 -0.264 0.792 0.9114 ENSG00000000460 57.9 0.0882 0.287 0.307 0.759 0.8955 ENSG00000000971 5817. -0.426 0.0883 -4.83 0.00000138 0.00001826 ENSG00000001036 1282. 0.241 0.0887 2.72 0.00658 0.0328
转换geneID
我们使用的MSigDB数据库的pathway 基因ID只有entrez和HGNC symbol两类,如果是ensemble id,需要转换
library(org.Hs.eg.db)library(tidyverse)ens2symbol <- AnnotationDbi::select(org.Hs.eg.db,key=res$row,columns="SYMBOL",keytype="ENSEMBL")ens2symbol <- as_tibble(ens2symbol)head(ens2symbol)
# A tibble: 6 x 2ENSEMBL SYMBOL<chr> <chr>1 ENSG00000000003 TSPAN62 ENSG00000000419 DPM13 ENSG00000000457 SCYL34 ENSG00000000460 C1orf1125 ENSG00000000971 CFH6 ENSG00000001036 FUCA2
- 合并数据;过滤NA值;去重;重复基因求stat(stat数据作为排序指标用于后续富集分析)
res2 <- inner_join(res, ens2symbol, by=c("row"="ENSEMBL")) %>%dplyr::select(SYMBOL, stat) %>%na.omit() %>%distinct() %>%group_by(SYMBOL) %>%summarize(stat=mean(stat))head(res2 )
# A tibble: 6 x 2SYMBOL stat<chr> <dbl>1 A1BG 0.6802 A1BG-AS1 -1.793 A2M -1.264 A2M-AS1 0.8755 A4GALT -4.146 A4GNT 0.00777
构建fgsea输入数据
- 基因排序值转换
library(fgsea)ranks <- deframe(res2)head(ranks, 20)
A1BG A1BG-AS1 A2M A2M-AS1 A4GALT A4GNT AAAS AACS0.679946437 -1.793291412 -1.259539478 0.875346116 -4.144839902 0.007772497 0.163986128 1.416071728AADACL4 AADAT AAGAB AAK1 AAMDC AAMP AAR2 AARS1-1.876311694 3.079128034 1.554279946 1.141522348 -2.147527241 -3.170612332 -2.364380163 4.495474603AARS2 AARSD1 AASDH AASDHPPT5.057470292 0.654208006 0.665531695 -0.353496148
- pathways的基因集合,上MSigDB下载基因集。演示使用KEGG基因集
pathways.hallmark <- gmtPathways("../../Result/GeneID/msigdb.v7.1.symbols_KEGG.gmt")pathways.hallmark %>%head() %>%lapply(head)
$KEGG_GLYCOLYSIS_GLUCONEOGENESIS[1] "ACSS2" "GCK" "PGK2" "PGK1" "PDHB" "PDHA1"$KEGG_CITRATE_CYCLE_TCA_CYCLE[1] "IDH3B" "DLST" "PCK2" "CS" "PDHB" "PCK1"$KEGG_PENTOSE_PHOSPHATE_PATHWAY[1] "RPE" "RPIA" "PGM2" "PGLS" "PRPS2" "FBP2"$KEGG_PENTOSE_AND_GLUCURONATE_INTERCONVERSIONS[1] "UGT1A10" "UGT1A8" "RPE" "UGT1A7" "UGT1A6" "UGT2B28"$KEGG_FRUCTOSE_AND_MANNOSE_METABOLISM[1] "MPI" "PMM2" "PMM1" "FBP2" "PFKM" "GMDS"$KEGG_GALACTOSE_METABOLISM[1] "GCK" "GALK1" "GLB1" "GALE" "B4GALT1" "PGM2"
- 运行
fgseaRes <- fgsea(pathways=pathways.hallmark, stats=ranks, nperm=1000)head(fgseaRes[order(pval), ])
- 从查看KEGG_REGULATION_OF_ACTIN_CYTOSKELETON富集分数分布
plotEnrichment(pathways.hallmark[["KEGG_REGULATION_OF_ACTIN_CYTOSKELETON"]],ranks) + labs(title="KEGG_REGULATION_OF_ACTIN_CYTOSKELETON")
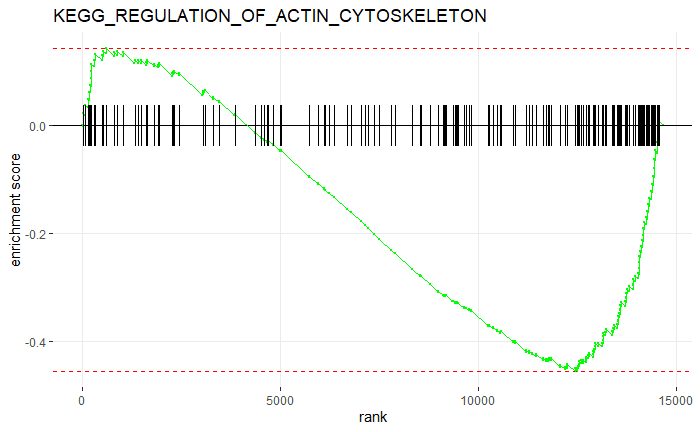
- 查看上下调通路结果
topPathwaysUp <- fgseaRes[ES > 0][head(order(pval), n=10), pathway]topPathwaysDown <- fgseaRes[ES < 0][head(order(pval), n=10), pathway]topPathways <- c(topPathwaysUp, rev(topPathwaysDown))plotGseaTable(pathways.hallmark[topPathways], ranks, fgseaRes,gseaParam=0.5)

- 其他展示方式
fgseaResTidy <- fgseaRes %>%as_tibble() %>%arrange(desc(NES))# Show in a nice table:fgseaResTidy %>%dplyr::select(-leadingEdge, -ES, -nMoreExtreme) %>%arrange(padj) %>%DT::datatable()ggplot(fgseaResTidy, aes(reorder(pathway, NES), NES)) +geom_col(aes(fill = padj<0.0001)) +coord_flip() +labs(x="Pathway", y="Normalized Enrichment Score",title="Hallmark pathways NES from GSEA") +theme_minimal()
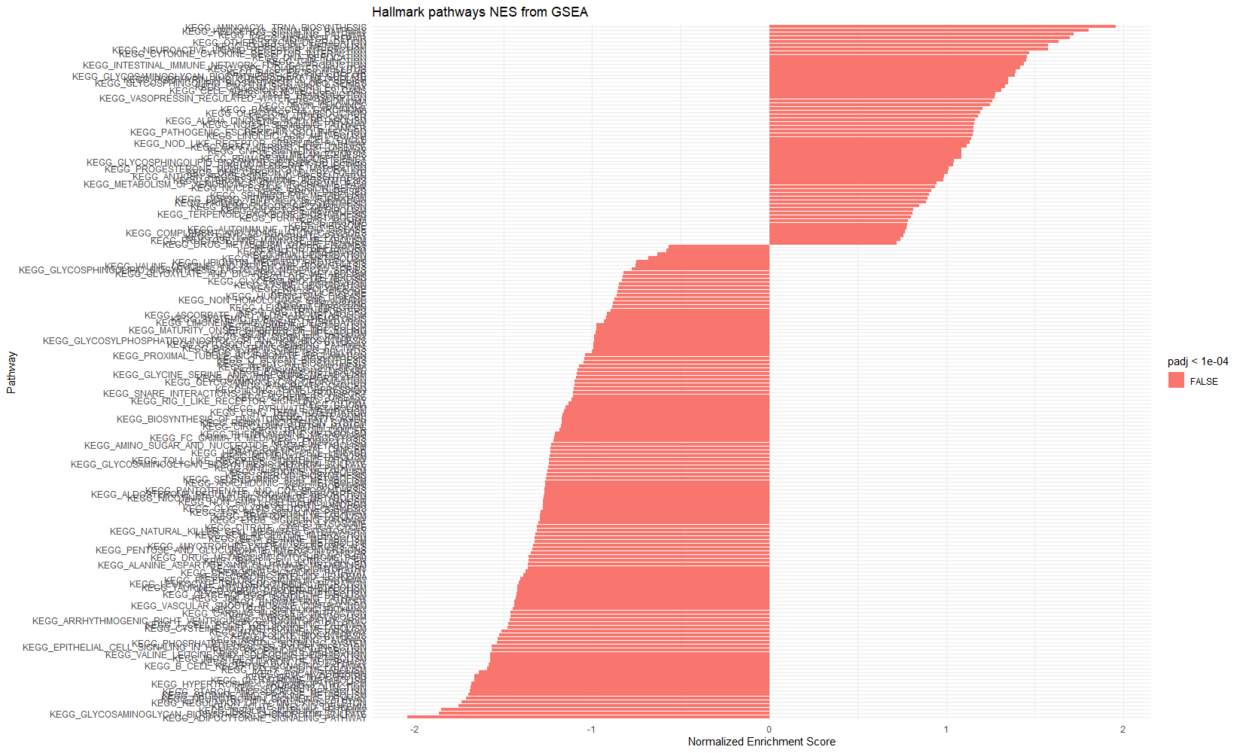
查看通路的基因
res_temp <- inner_join(res, ens2symbol, by=c("row"="ENSEMBL"))pathways.hallmark %>%enframe("pathway", "SYMBOL") %>%unnest(cols = c(SYMBOL)) %>%inner_join(res_temp , by="SYMBOL") %>%head()
# A tibble: 6 x 9pathway SYMBOL row baseMean log2FoldChange lfcSE stat pvalue padj<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>1 KEGG_GLYCOLYSIS_GLUCONEOGENESIS ACSS2 ENSG00000131069 669. -0.269 0.114 -2.35 0.0188 0.07562 KEGG_GLYCOLYSIS_GLUCONEOGENESIS GCK ENSG00000106633 28.8 0.305 0.374 0.815 0.415 0.6623 KEGG_GLYCOLYSIS_GLUCONEOGENESIS PGK1 ENSG00000102144 7879. -0.300 0.353 -0.850 0.395 0.6424 KEGG_GLYCOLYSIS_GLUCONEOGENESIS PDHB ENSG00000168291 648. -0.257 0.102 -2.52 0.0117 0.05215 KEGG_GLYCOLYSIS_GLUCONEOGENESIS PDHA1 ENSG00000131828 651. -0.0744 0.104 -0.715 0.475 0.7106 KEGG_GLYCOLYSIS_GLUCONEOGENESIS PGM2 ENSG00000169299 302. -0.315 0.136 -2.33 0.0201 0.0797
其他用法
- miR targets
fgsea(pathways=gmtPathways("msigdb/c3.mir.v6.2.symbols.gmt"), ranks, nperm=1000) %>%as_tibble() %>%arrange(padj)
- GO annotations
fgsea(pathways=gmtPathways("msigdb/c5.all.v6.2.symbols.gmt"), ranks, nperm=1000) %>%as_tibble() %>%arrange(padj)
- 非人物种
library(biomaRt)mart <- useDataset("mmusculus_gene_ensembl", mart=useMart("ensembl"))bm <- getBM(attributes=c("ensembl_gene_id", "hsapiens_homolog_associated_gene_name"), mart=mart) %>%distinct() %>%as_tibble() %>%na_if("") %>%na.omit()bm
参考
参考文章如引起任何侵权问题,可以与我联系,谢谢。

