from nd2reader import ND2Readerfrom cane import cane_3dimport imageioimport numpy as npimport timeimport matplotlib.pyplot as pltimport napari## Nikon data to numpy arrayimages = ND2Reader('/mnt/data/CA3-CA1-tracing.nd2')ny,nx = images.frame_shapenz = len(images.metadata['z_levels'])Img = np.zeros((nz,ny,nx),dtype='float64')for i in range(0,nz):Img[i,:,:] = images[i]#viewer = napari.view_image(Img)
nikon nd2 format metedata information
All metadata properties are:
width: the width of the image in pixelsheight: the height of the image in pixelsdate: the date the image was takenfields_of_view: the fields of view in the imageframes: a list of all frame numbersz_levels: the z levels in the imagetotal_images_per_channel: the number of images per color channelchannels: the color channelspixel_microns: the amount of microns per pixelrois: the regions of interest (ROIs) defined by the userexperiment: information about the nature and timings of the ND experiment
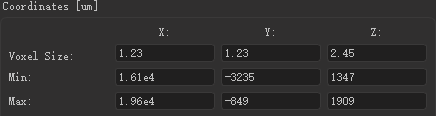
高灿灿拍摄的神经元像素尺寸大小: