基本概念
过拟合
在模型学习数据普遍化模式的过程中,它还学习了每个样本特有的噪音特征,这样的模型称为过拟合。
过拟合的模型可能在原数据集上的表现非常好,但是泛化能力很差,也就是换一个数据集表现就很差,这就是由于过拟合导致的。
模型调优
- 几乎所有的预测模型方法都含有调优参数,用现有的数据来调整这些参数,从而给出最好的预测,这个过程称为模型调优。
数据分割
数据量较小时应避免划分测试集。
如果某类的样本量明显少于其他类,那么简单的随机划分会导致训练集和测试集结果大相径庭,应使用分层随机抽样法
重抽样技术
- K折交叉验证
- 广义交叉验证
- 重复训练/测试集划分
- Bootstrap方法
数据划分建议
- 样本量较少,笔者建议使用10折交叉验证
- 如果目标不是得到最好的模型表现的估计,而是在几个不同的模型中进行选择,那么最好使用Bootstrap方法
计算
## 加载R包和数据library(AppliedPredictiveModeling)data(twoClassData)str(predictors)## 'data.frame': 208 obs. of 2 variables:## $ PredictorA: num 0.158 0.655 0.706 0.199 0.395 ...## $ PredictorB: num 0.1609 0.4918 0.6333 0.0881 0.4152 ...str(classes)## Factor w/ 2 levels "Class1","Class2": 2 2 2 2 2 2 2 2 2 2 ...set.seed(1)
# 划分数据集library(caret)## 载入需要的程辑包:lattice## 载入需要的程辑包:ggplot2trainingRows <- createDataPartition(classes, p = 0.8, list = F) # 也可进行分层抽样head(trainingRows)## Resample1## [1,] 1## [2,] 2## [3,] 3## [4,] 7## [5,] 8## [6,] 9
变为训练集和测试集:
trainPredictors <- predictors[trainingRows, ]trainClasses <- classes[trainingRows]testPredictors <- predictors[-trainingRows, ]testClasses <- classes[-trainingRows]str(trainPredictors)## 'data.frame': 167 obs. of 2 variables:## $ PredictorA: num 0.1582 0.6552 0.706 0.0658 0.3086 ...## $ PredictorB: num 0.161 0.492 0.633 0.179 0.28 ...str(testPredictors)## 'data.frame': 41 obs. of 2 variables:## $ PredictorA: num 0.1992 0.3952 0.425 0.0847 0.2909 ...## $ PredictorB: num 0.0881 0.4152 0.2988 0.0548 0.3021 ...
## 重抽样set.seed(1)repeatedSplits <- createDataPartition(trainClasses, p = 0.8, times = 3)str(repeatedSplits)## List of 3## $ Resample1: int [1:135] 1 2 3 4 6 7 9 10 11 12 ...## $ Resample2: int [1:135] 1 2 3 4 5 6 7 9 10 11 ...## $ Resample3: int [1:135] 1 2 3 4 5 7 8 9 11 12 ...
## K折交叉验证set.seed(1)cvSplits <- createFolds(trainClasses, k = 10, returnTrain = T)str(cvSplits)## List of 10## $ Fold01: int [1:150] 1 2 4 5 6 7 8 10 11 13 ...## $ Fold02: int [1:150] 1 2 3 4 6 7 8 9 10 11 ...## $ Fold03: int [1:150] 1 3 4 5 6 7 8 9 10 11 ...## $ Fold04: int [1:150] 1 2 3 4 5 6 7 8 9 10 ...## $ Fold05: int [1:150] 2 3 4 5 6 7 8 9 10 11 ...## $ Fold06: int [1:150] 1 2 3 4 5 6 7 8 9 11 ...## $ Fold07: int [1:150] 1 2 3 4 5 6 7 9 10 12 ...## $ Fold08: int [1:151] 1 2 3 4 5 6 8 9 10 11 ...## $ Fold09: int [1:151] 1 2 3 5 6 7 8 9 10 11 ...## $ Fold10: int [1:151] 1 2 3 4 5 7 8 9 10 11 ...fold1 <- cvSplits[[1]] # 第一折的行号cvPredictors1 <- trainPredictors[fold1, ] # 得到第一份90%的样本cvClass1 <- trainClasses[fold1]nrow(trainPredictors)## [1] 167nrow(cvPredictors1)## [1] 150
## R基础建模## 训练trainPredictors <- as.matrix(trainPredictors)knnFit <- knn3(x = trainPredictors, y = trainClasses, k = 5)knnFit## 5-nearest neighbor model## Training set outcome distribution:#### Class1 Class2## 89 78## 预测testPredictions <- predict(knnFit, newdata = testPredictors, type = "class")head(testPredictions)## [1] Class2 Class1 Class1 Class2 Class1 Class2## Levels: Class1 Class2str(testPredictions)## Factor w/ 2 levels "Class1","Class2": 2 1 1 2 1 2 2 1 2 2 ...
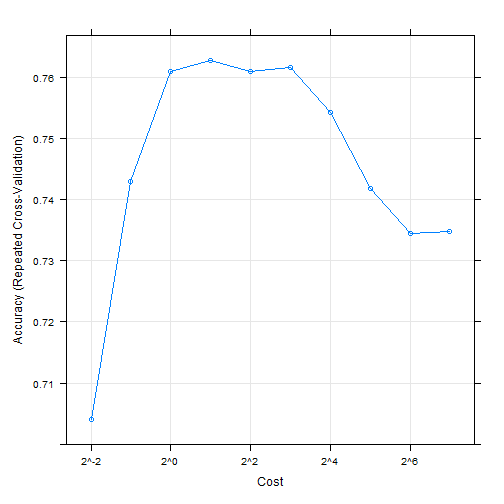
## 决定调优参数library(caret)data("GermanCredit")set.seed(1056)svmFit <- train(Class ~.,data = GermanCredit,method = "svmRadial")## 进行预处理,并使用重复5折交叉验证set.seed(1056)svmfit <- train(Class ~.,data = GermanCredit,method = "svmRadial",preProc = c("center" ,"scale"),tuneLength = 10,trControl = trainControl(method = "repeatedcv", repeats = 5)) # 其实这个函数我感觉比现在的tidymodels和mlr3的写法都要简洁...svmfit## Support Vector Machines with Radial Basis Function Kernel#### 1000 samples## 61 predictor## 2 classes: 'Bad', 'Good'#### Pre-processing: centered (61), scaled (61)## Resampling: Cross-Validated (10 fold, repeated 5 times)## Summary of sample sizes: 900, 900, 900, 900, 900, 900, ...## Resampling results across tuning parameters:#### C Accuracy Kappa## 0.25 0.7040 0.01934723## 0.50 0.7430 0.24527603## 1.00 0.7610 0.35046362## 2.00 0.7628 0.38285072## 4.00 0.7610 0.39239970## 8.00 0.7616 0.40357861## 16.00 0.7542 0.39860268## 32.00 0.7418 0.37677389## 64.00 0.7344 0.36165095## 128.00 0.7348 0.36361822#### Tuning parameter 'sigma' was held constant at a value of 0.009718427## Accuracy was used to select the optimal model using the largest value.## The final values used for the model were sigma = 0.009718427 and C = 2.plot(svmfit, scales = list(x=list(log = 2)))
## 比较模型set.seed(1056)logistic <- train(Class ~.,data = GermanCredit,method = "glm",trControl = trainControl(method = "repeatedcv", repeats = 5))logistic## Generalized Linear Model#### 1000 samples## 61 predictor## 2 classes: 'Bad', 'Good'#### No pre-processing## Resampling: Cross-Validated (10 fold, repeated 5 times)## Summary of sample sizes: 900, 900, 900, 900, 900, 900, ...## Resampling results:#### Accuracy Kappa## 0.749 0.3661277resamp <- resamples(list(svm = svmfit, logi = logistic))summary(resamp)#### Call:## summary.resamples(object = resamp)#### Models: svm, logi## Number of resamples: 50#### Accuracy## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's## svm 0.69 0.7425 0.77 0.7628 0.7800 0.84 0## logi 0.65 0.7200 0.75 0.7490 0.7775 0.88 0#### Kappa## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's## svm 0.1944444 0.3385694 0.3882979 0.3828507 0.4293478 0.5959596 0## logi 0.1581633 0.2993889 0.3779762 0.3661277 0.4240132 0.7029703 0summary(diff(resamp))#### Call:## summary.diff.resamples(object = diff(resamp))#### p-value adjustment: bonferroni## Upper diagonal: estimates of the difference## Lower diagonal: p-value for H0: difference = 0#### Accuracy## svm logi## svm 0.0138## logi 0.0002436#### Kappa## svm logi## svm 0.01672## logi 0.07449

