3.1 导读
单细胞降维聚类之后需要重要的几个步骤
- 自定义marker基因 可视化
- 借助Marker基因,定型细胞类型
- 细胞比例的差异分析
3.2 自定义Marker基因可视化
定义maker基因list
# 参考文献https://www.nature.com/articles/s41591-018-0096-5#Ack1maker_gene = {"endothelial_cells":[ "CLDN18", "FOLR1", "AQP4" , "PEBP4"],"endothilial":["CLDN5", "FLT1", "CDH5 ","RAMP2"],"epithelial_cells": ["CAPS", "TMEM190", "PIFO","SNTN"],"fibroblasts":["COL1A1", "DCNCOL1A2","C1R9"] ,"B_cells":["CD79A","IGKC", "IGLC3","IGHG3","LYZ", "MARCO", "CD68" ,"CD16a", "FCGR3A"],"T_cells":["CD3D","TRAC", "TRBC1" , "TRBC"]}
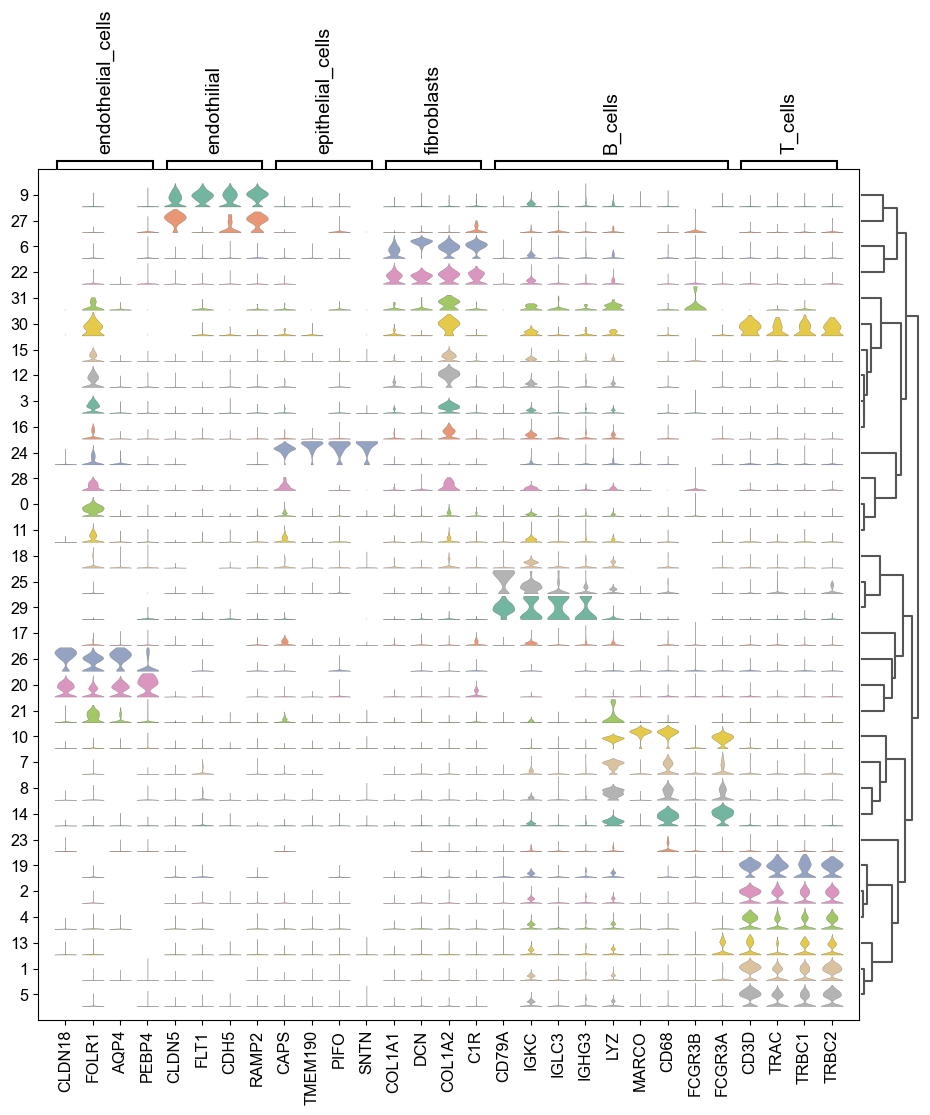
堆积小提琴图
sc.pl.stacked_violin(adata_all,maker_gene, groupby="louvain_2",use_raw=False, dendrogram=True, standard_scale ='var',row_palette = 'Set2',scale='count')
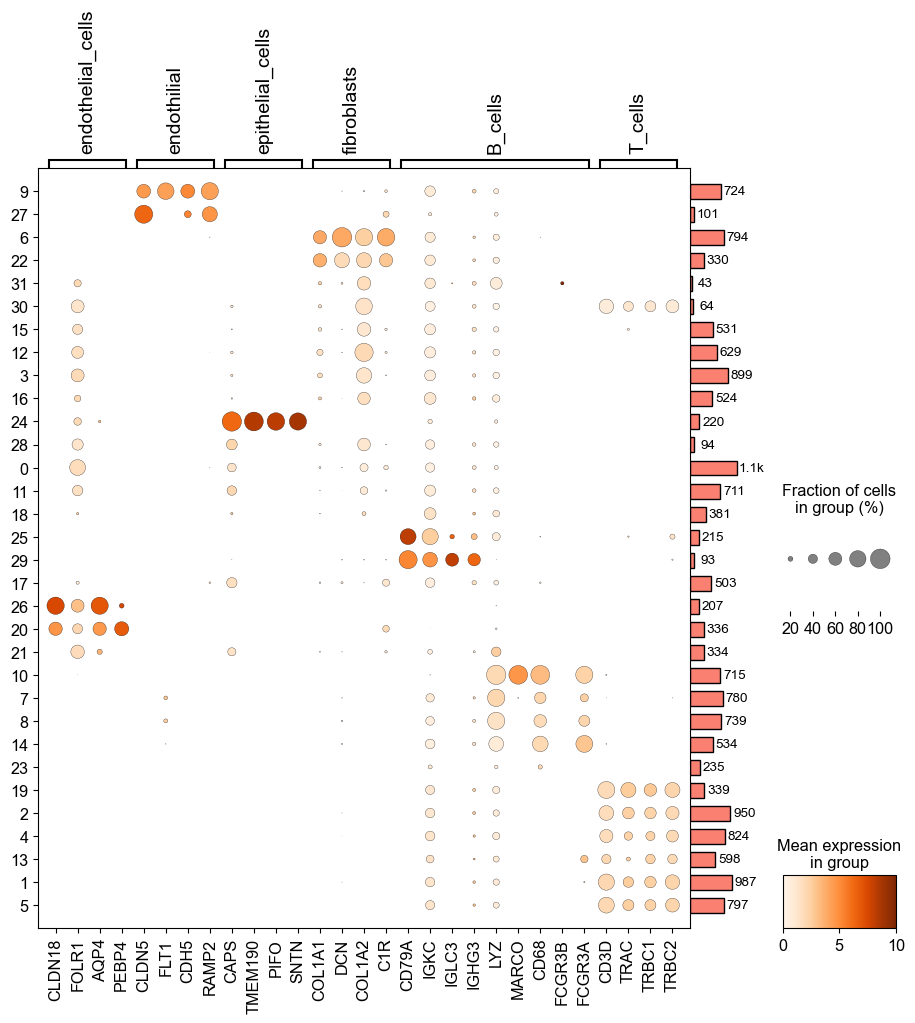
气泡图
sc.settings.set_figure_params(dpi=50, facecolor='white', dpi_save =100)myplot = sc.pl.dotplot(adata_all, maker_gene, groupby='louvain_2',dendrogram=True,color_map='Oranges', mean_only_expressed=True,return_fig=True,dot_max=1,dot_min=0.05,figsize=(11,10))myplot.add_totals().show()
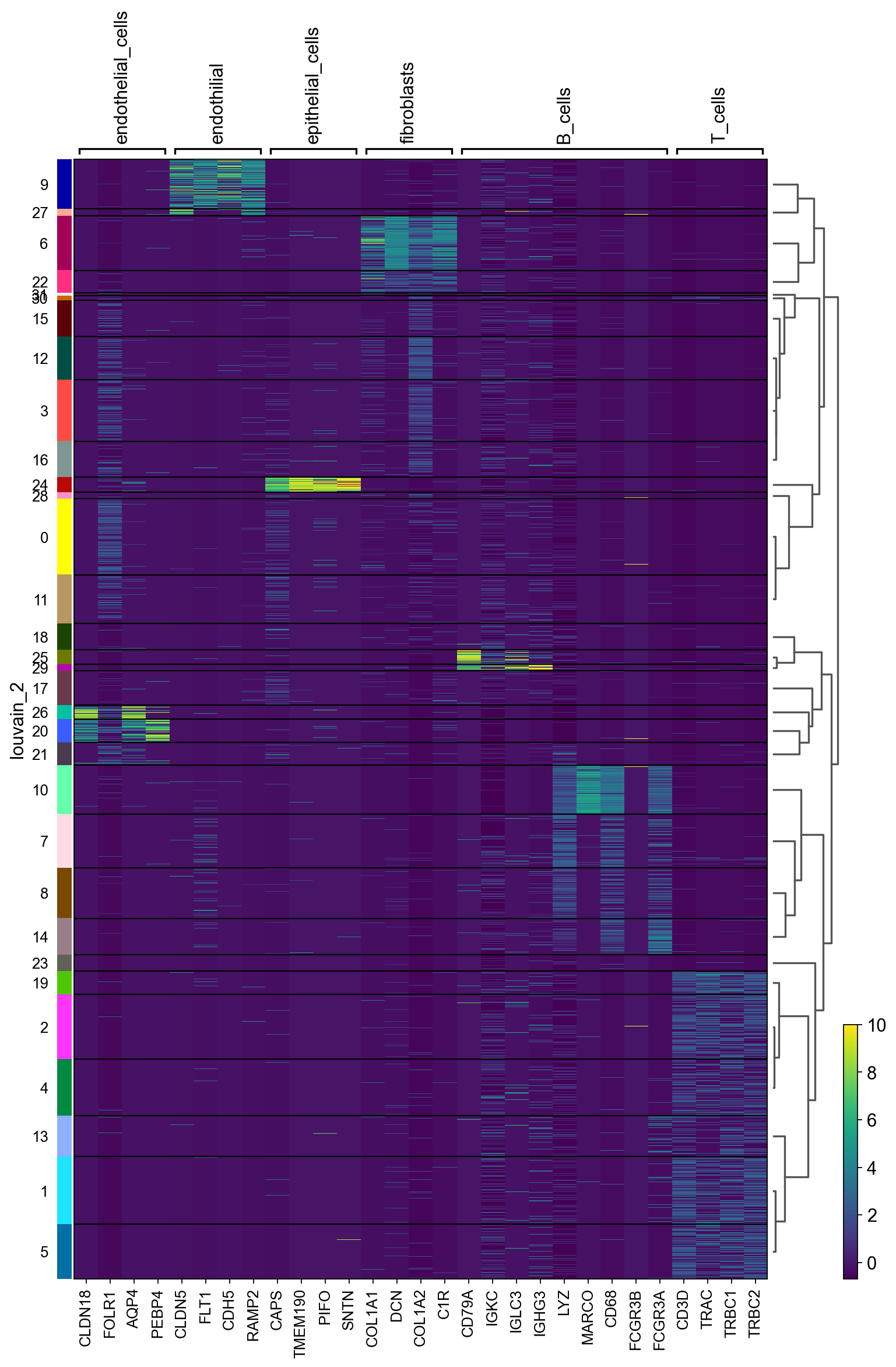
聚类热图
sc.pl.heatmap(adata_all, maker_gene, groupby='louvain_2',dendrogram=True,figsize=(11,16))
可调节figsize 显示整体图形大小
3.3 未知Marker基因定型
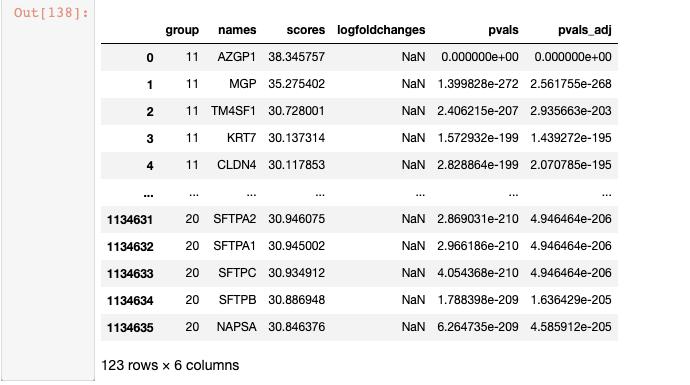
取出所有cluster中 Marker基因top=5 进行去重。然后可视化小提琴图或者点图
df_markergene_all = sc.get.rank_genes_groups_df(adata_all, group=list(adata_all.obs.louvain_2.unique()),key= "louvain_2_rankgenes")marke_list = list(df_markergene_all.groupby(['group']).head(5).drop_duplicates(subset='names')['names'])
可视化画图
sc.settings.set_figure_params(dpi=50, facecolor='white', dpi_save =100)sc.pl.stacked_violin(adata_all,maker_list, groupby="louvain_2",use_raw=False, dendrogram=True, standard_scale ='var',row_palette = 'Set2',scale='count')
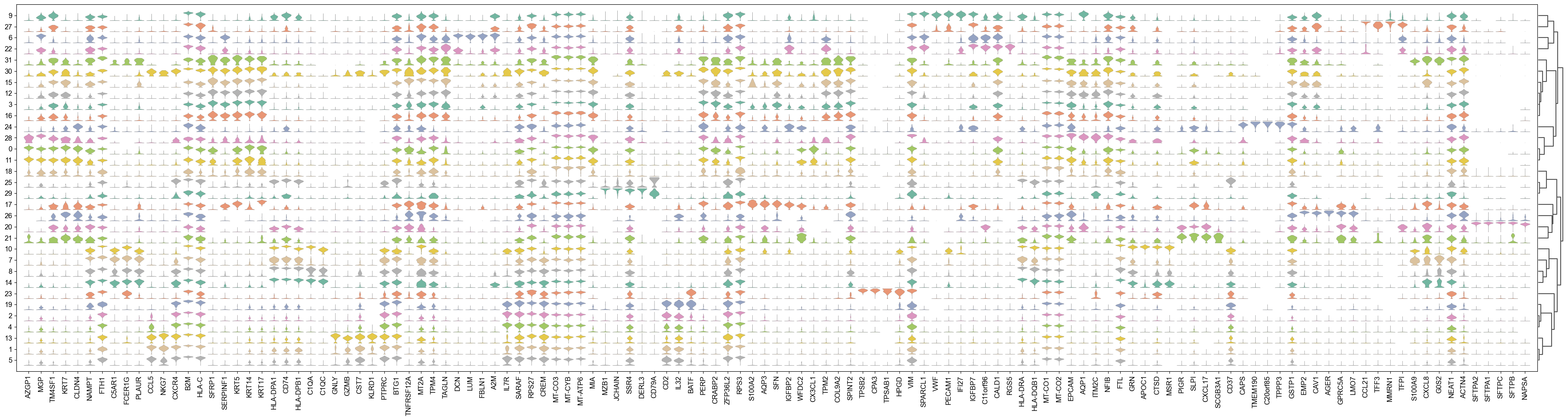
查找相关基因进行定型 如果找不到相关信息,后续通过GO/KEGG 富集进行定位基因功能。后续会有相关分析。
定型细胞注释
通过自定义细胞已知细胞类型定义,如果想定义更详细的细胞类型可查找相关的Marker 进行注释。大体分为一下格式
cell_type = {"T_cells":[5,1,13,4,2,19],"B_cells":[14,8,7,10,29,25,18,23],"endothelial_cells":[20,26,21,0,11],"fibroblasts":[28,6,22,31,30,15,12,3,16,28],"endothilial":[9,27],"epithelial_cells":[24,17]}adata_all.obs['cell_type'] = adata_all.obs['louvain_2'].astype(str) ## 赋值类型for key in cell_type:mask = adata_all.obs['cell_type'].isin(map(str,cell_type[key]))adata_all.obs.loc[mask,"cell_type"] = key
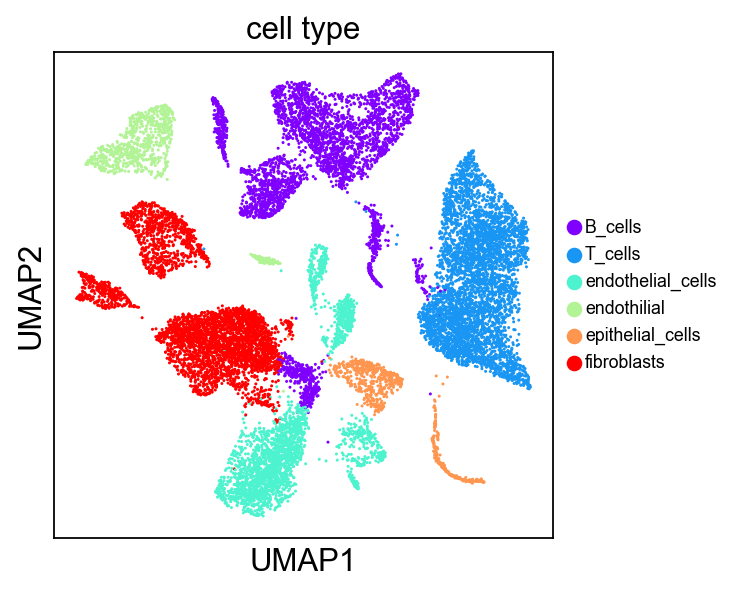
可视化umap 按照新的赋值
sc.settings.set_figure_params(dpi=80, facecolor='white', dpi_save =100)sc.pl.umap(adata_all, title="cell type", color = "cell_type", legend_fontsize = 8,frameon = True, wspace =0.3, color_map='Blues', palette='rainbow', use_raw =False)
总结
细胞定型是一个技术活。需要读取文献进行注释分析,细胞定型完毕。后续的分析就可以简单进行了。


