一步运行
data_hyper <- runHyper(data_deg,dir = "hyper_out", GO = TRUE, KEGG = TRUE)
分解动作
富集分析运算
## step only resolvedata_hyper <- hyperResolve( data_deg, GO = FALSE, KEGG = TRUE)
结果输出
## step summary resultshyperSummary( data_hyper, dir = "hyper_out", prefix = "3-runHyper", top = 10)
衍生工具
enrichGO2(gene,TERM2GENE,TERM2NAME = NA,organism = "UNKNOW",keyType = "SYMBOL",ont = "ALL",pvalueCutoff = 0.05,pAdjustMethod = "BH",universe,qvalueCutoff = 0.2,minGSSize = 10,maxGSSize = 500,pool = FALSE)
可视化
适用于GO的柱状图
gob <- hyperRes(data_hyper)[["hyperGO_res"]][["limma"]][["Up"]]p_hypergbar <- enrichBar(gob, top = 10, plot_title = "limma")ggplot2::ggsave(p_hypergbar, filename = "hyper_go_bar.pdf", width = 3000, height = 3600, units = "px", limitsize = FALSE, device = cairo_pdf, dpi = 300)
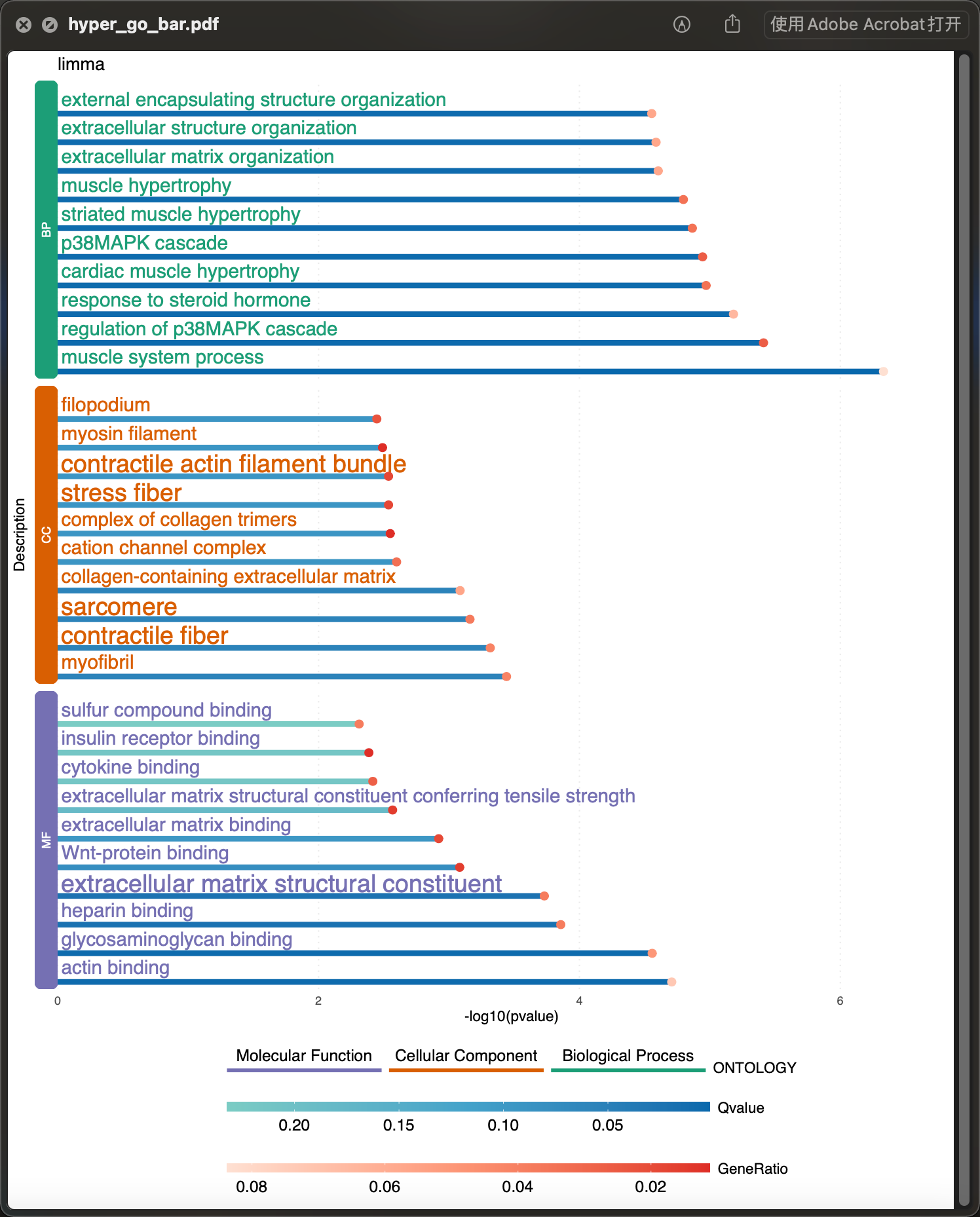
通用柱状图
eob <- hyperRes(data_hyper)[["hyperKEGG_res"]][["limma"]][["Up"]]p_hyperebar <- hyperBar(eob, top = 10)ggplot2::ggsave(p_hyperbar, filename = "hyper_kegg_bar.pdf", width = 2100, height = 2400, units = "px", limitsize = FALSE, device = cairo_pdf)
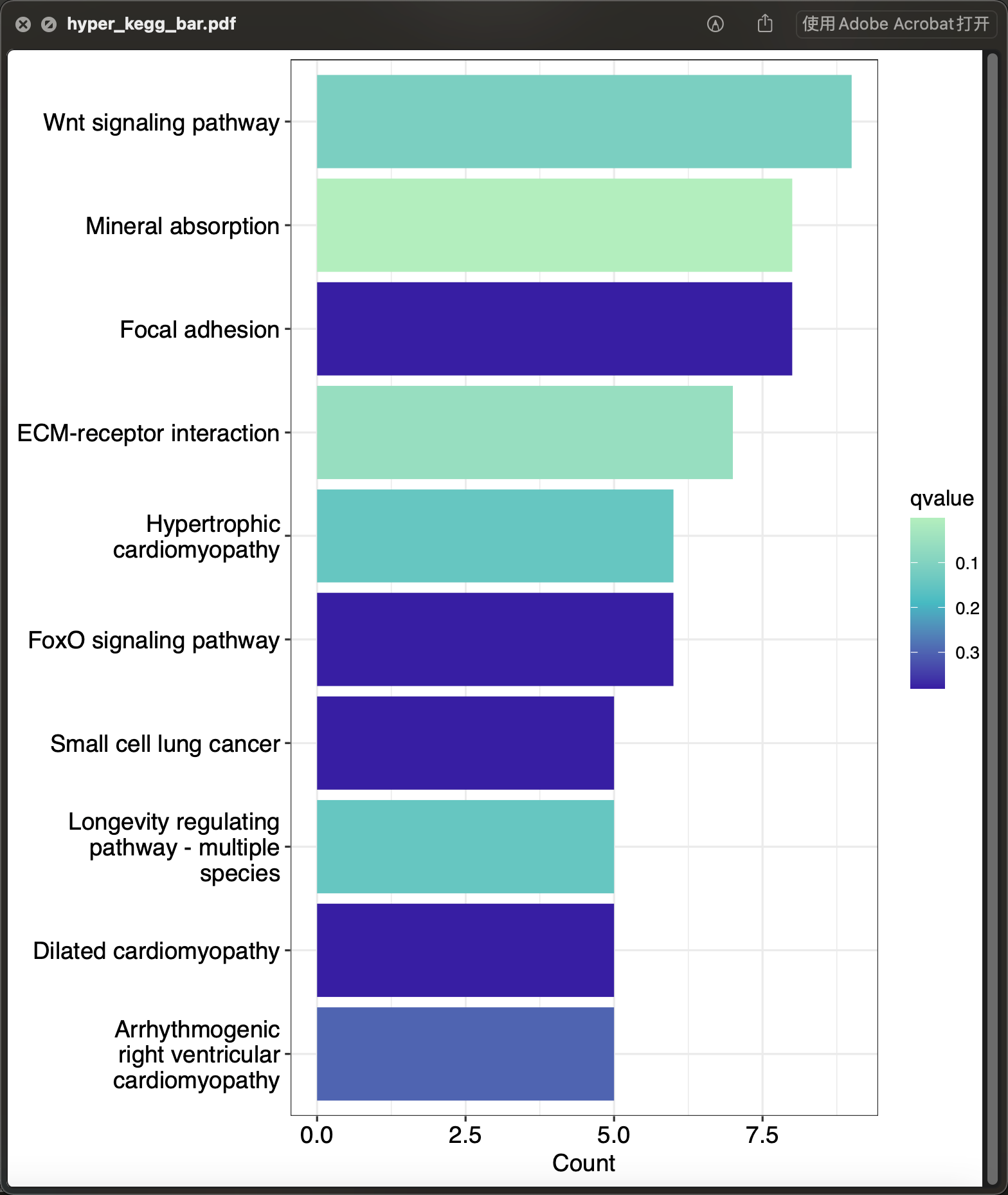
圈图
## how many Common GO IDs among limma edgeR EDSeq2 mergedat_d <- modelEnrich(data_hyper, dataBase = "KEGG", orderBy = "pvalue", head = 10)[["diff"]]cc_file_name <- "KEGG_compareEnrichCircle.pdf"compareEnrichCircle(result_g = dat_d, filename = cc_file_name, mar = c(8, 0, 0, 19))





