一步运行
## complete GSEA moduledata_gse <- runGSEA(data_hyper,dir = "gse_out", GO = TRUE, KEGG = TRUE)
分步运行
计算
## step only resolvedata_gse <- gseResolve( data_hyper, GO = FALSE, KEGG = TRUE)
结果整理
## step summary resultsgseSummary( data_gse, dir = "gse_out", prefix = "4-runGSEA", top = 10)
辅助工具
自定义GO注释的富集分析
gseGO2( geneList, ont = "ALL", TERM2GENE, TERM2NAME = NA, organism = "UNKNOW", keyType = "SYMBOL", exponent = 1, minGSSize = 10, maxGSSize = 500, eps = 1e-10, pvalueCutoff = 0.05, pAdjustMethod = "BH", verbose = TRUE, seed = FALSE, by = "fgsea", ...)
可视化工具
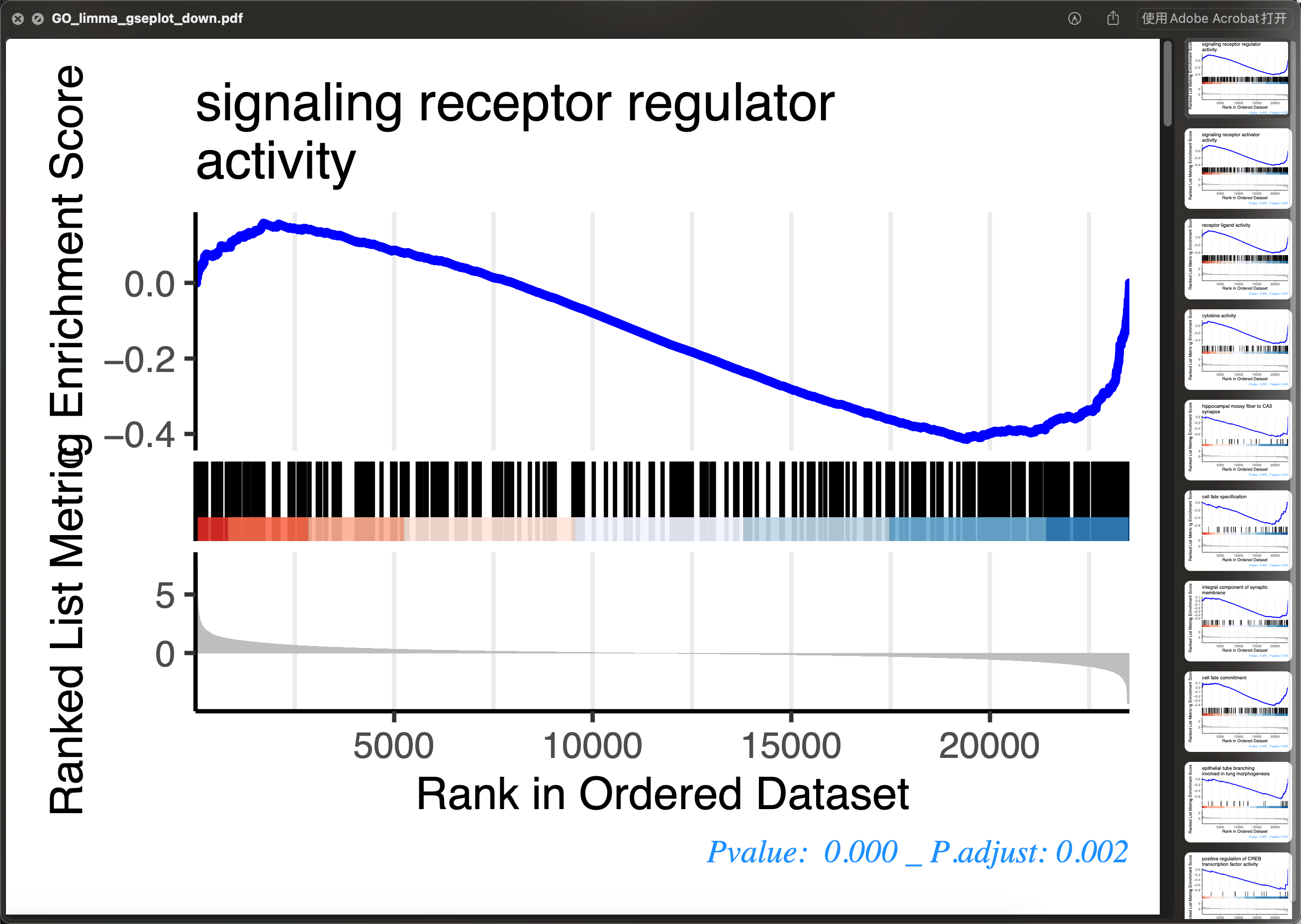
gsea 图
p_gsea_list <- GSEAplot(gsob,top = 10)down_plots_pdf <- "GO_limma_gseplot_down.pdf"pdf(down_plots_pdf,height = 3,width = 4)invisible(lapply(p_gsea_list[["down_plots"]], print))dev.off()
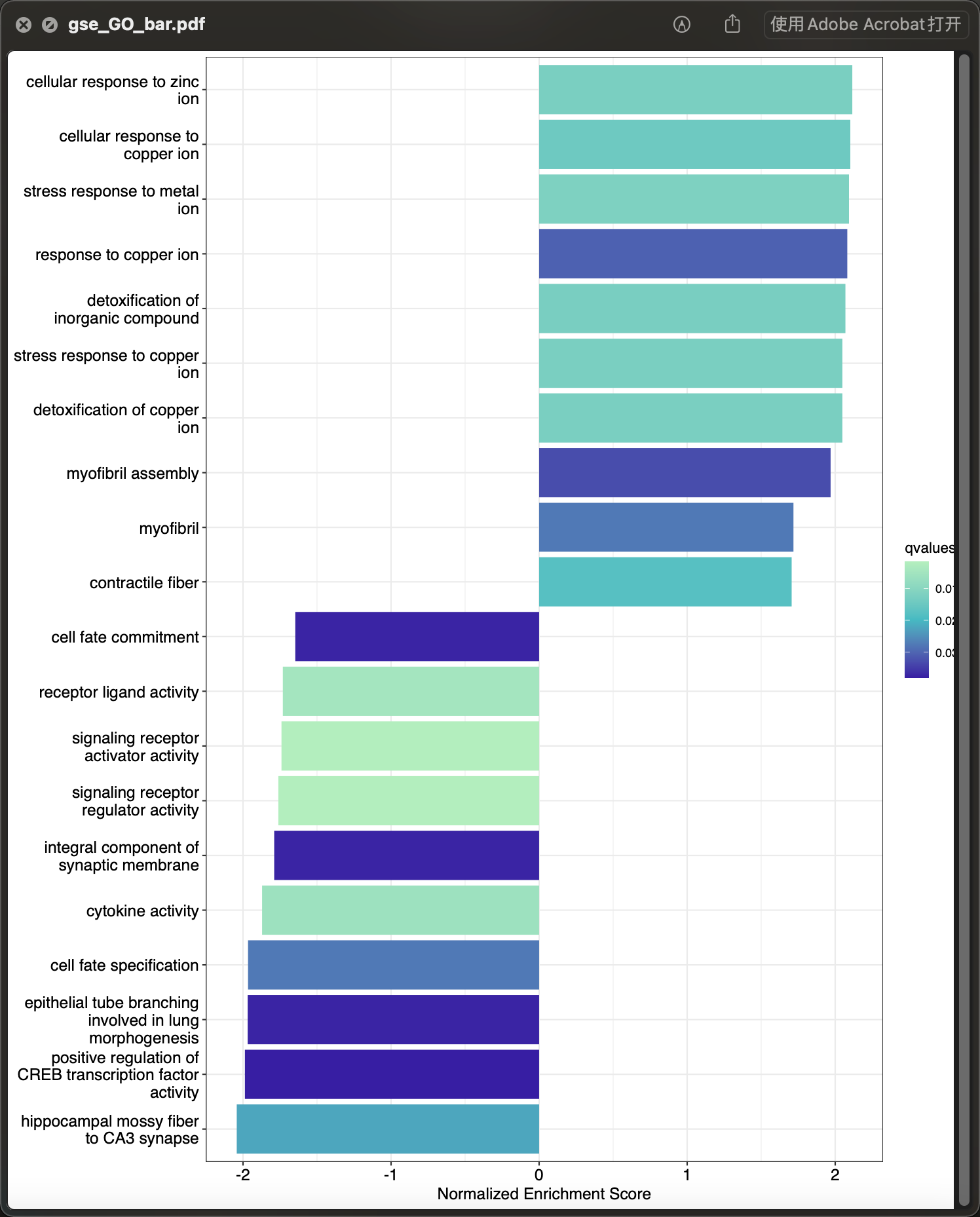
gse 柱状图
gsob <- gseRes(data_gse)[["gseGO_res"]][["limma"]]p_gsegbar <- GSEAbar(gsob,top = 10)ggplot2::ggsave(p_gsegbar,filename = "gse_GO_bar.pdf", width = 3000,height = 3600,units = "px",limitsize = FALSE,device = cairo_pdf)