导入库
from rdkit import Chemfrom rdkit.Chem import Drawfrom rdkit.Chem.Draw import SimilarityMapsfrom IPython.display import SVGimport numpy as npimport rdkitprint(rdkit.__version__)

2020.09.1
基于Morgan指纹的相似图绘制
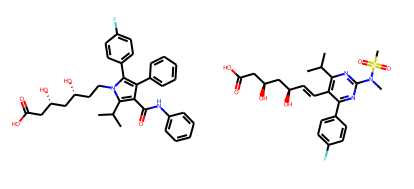
atorvastatin = Chem.MolFromSmiles('O=C(O)C[C@H](O)C[C@H](O)CCn2c(c(c(c2c1ccc(F)cc1)c3ccccc3)C(=O)Nc4ccccc4)C(C)C')rosuvastatin = Chem.MolFromSmiles('OC(=O)C[C@H](O)C[C@H](O)\C=C\c1c(C(C)C)nc(N(C)S(=O)(=O)C)nc1c2ccc(F)cc2')Draw.MolsToGridImage((atorvastatin,rosuvastatin))


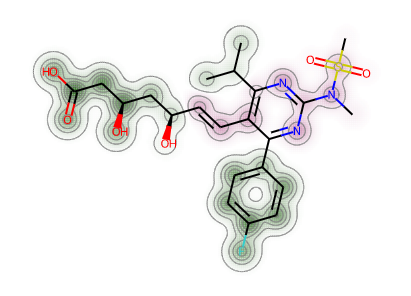
d = Draw.MolDraw2DSVG(400, 400)_, maxWeight = SimilarityMaps.GetSimilarityMapForFingerprint(atorvastatin, rosuvastatin,lambda m, i: SimilarityMaps.GetMorganFingerprint(m, i, radius=2, fpType='bv'),draw2d=d)d.FinishDrawing()SVG(d.GetDrawingText())


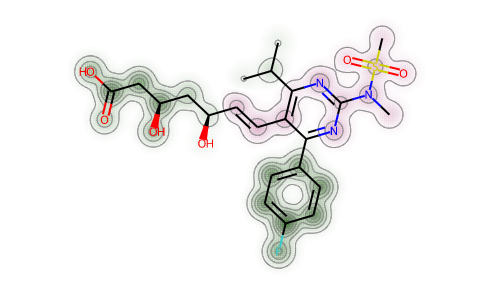
d = Draw.MolDraw2DSVG(400, 400)_, maxWeight = SimilarityMaps.GetSimilarityMapForFingerprint(atorvastatin, rosuvastatin,lambda m, i: SimilarityMaps.GetMorganFingerprint(m, i, radius=2, fpType='count'),draw2d=d)d.FinishDrawing()SVG(d.GetDrawingText())


from rdkit.Chem import rdMolDescriptorsator_contribs = rdMolDescriptors._CalcCrippenContribs(atorvastatin)d = Draw.MolDraw2DSVG(400, 400)SimilarityMaps.GetSimilarityMapFromWeights(atorvastatin,[x[0] for x in ator_contribs],draw2d=d)d.FinishDrawing()SVG(d.GetDrawingText())

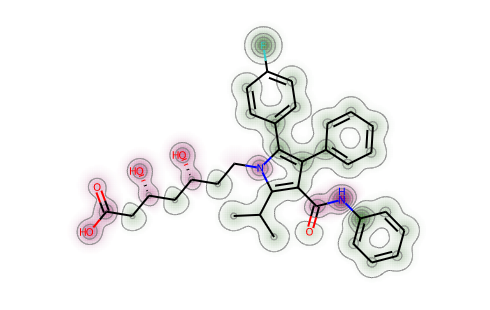

from rdkit.Chem import rdEHTToolsfrom rdkit.Chem import rdDistGeommh = Chem.AddHs(atorvastatin)rdDistGeom.EmbedMolecule(mh)_,res = rdEHTTools.RunMol(mh)static_chgs = res.GetAtomicCharges()[:atorvastatin.GetNumAtoms()]d = Draw.MolDraw2DSVG(400, 400)SimilarityMaps.GetSimilarityMapFromWeights(atorvastatin,list(static_chgs),draw2d=d)d.FinishDrawing()SVG(d.GetDrawingText())

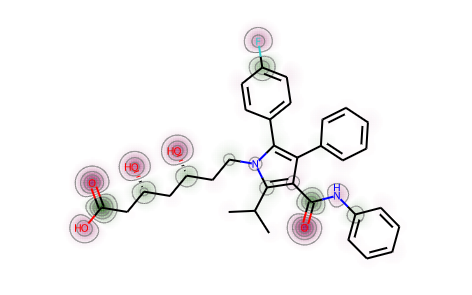

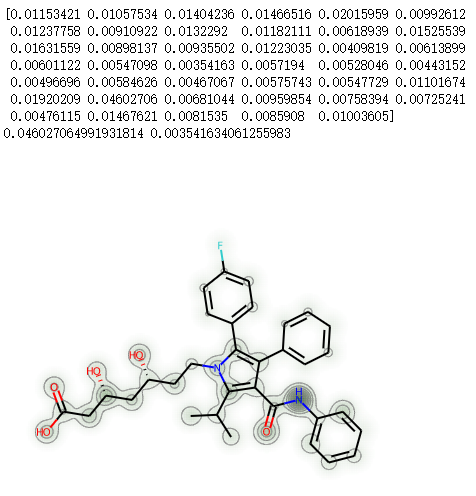
mh = Chem.AddHs(atorvastatin)ps = rdDistGeom.ETKDGv2()ps.pruneRmsThresh = 0.5ps.randomSeed = 0xf00drdDistGeom.EmbedMultipleConfs(mh,10,ps)print(f'Found {mh.GetNumConformers()} conformers')chgs = []for conf in mh.GetConformers():_,res = rdEHTTools.RunMol(mh,confId=conf.GetId())chgs.append(res.GetAtomicCharges()[:atorvastatin.GetNumAtoms()])chgs = np.array(chgs)mean_chgs = np.mean(chgs,axis=0)std_chgs = np.std(chgs,axis=0)d = Draw.MolDraw2DSVG(400, 400)SimilarityMaps.GetSimilarityMapFromWeights(atorvastatin,list(mean_chgs),draw2d=d)d.FinishDrawing()SVG(d.GetDrawingText())

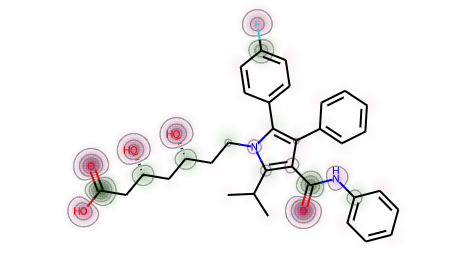

print(std_chgs)print(max(std_chgs),min(std_chgs))d = Draw.MolDraw2DSVG(400, 400)SimilarityMaps.GetSimilarityMapFromWeights(atorvastatin,list(std_chgs),draw2d=d)d.FinishDrawing()SVG(d.GetDrawingText())