介绍:https://vdjtools-doc.readthedocs.io/en/master/
1.下载
对于linux服务器而言,可以直接通过https://github.com/mikessh/vdjtools/releases/tag/1.2.1,进行下载。
zip安装包中包含了所需的jar文件,无需编译,下载即用。
安装相关的R包:
java -jar vdjtools.jar Rinstall
2.使用
事例:
# -p for plotting, -f specifies metadata column for coloring,# -n tells that factor is continuous$VDJTOOLS CalcSegmentUsage -m metadata.txt -p -f age -n out/2
输入:
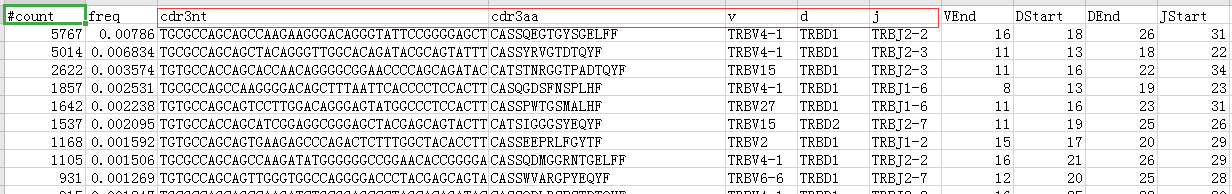
克隆丰度表,对于10x免疫组库的结果而言,可以使用获得的filtered_contig_annotations.csv
对于VDJtools而言,所需的clonotype数据包括这些:
- Variable (V) segment name. V区基因名
- Diversity (D) segment name for some of the receptor chains (TRB, TRD and IGH). Set to . if not aplicable or D segment was not identified. D区基因名,如果是.则是未获得
- Joining (J) segment name. J区基因名
- Complementarity determining region 3 nucleotide sequence (CDR3nt). CDR3 starts with Variable region reference point (conserved Cys residue) and ends with Joining segment reference point (conserved PheTrp). CDR3的核苷酸序列
- Translated CDR3 sequence (CDR3aa). CDR3的氨基酸序列
- Somatic hypermutations (SHMs) in the variable segment (antibody only, planned). V区的体细胞突变
格式转换:
将10x的结果数据转换成需要的格式
10x数据:
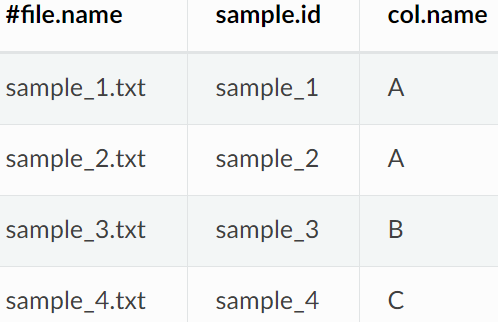
将样本处理成需要的格式:
Metadata数据格式如下:
分析模块
基础模块:
汇总统计
- CalcBasicStats Computes summary statistics for samples: read counts, mean clonotype sizes, number of non-functional clonotypes, etc
- CalcSegmentUsage Computes Variable (V) and Joining (J) segment usage profiles
- CalcSpectratype Computes spectratype, the distribution of clonotype abundance by CDR3 sequence length
- PlotFancySpectratype Plots spectratype explicitly showing top N clonotypes
- PlotFancyVJUsage Plots the frequency of different V-J pairings
- PlotSpectratypeV Plots distribution of V segment abundance by resulting CDR3 sequence length
多样性评估
丰富和多样性
- PlotQuantileStats Visualizes repertoire clonality
- RarefactionPlot Performs rarefaction analysis
- CalcDiversityStats Computes repertoire diversity estimates
样本间overlap
样本间共享的克隆型
- OverlapPair Computes intersection between a pair of samples
- CalcPairwiseDistances Computes pairwise intersections for a list of samples
- ClusterSamples Performs sample clusterization according to the results of batch intersection
- TrackClonotypes Time-course analysis for a sequence of samples
Pre-processing
过滤和重采样
- Correct Performs a frequency-based erroneous clonotype correction
- Decontaminate Filters possible cross-sample contaminations in a set of samples
- DownSample Performs down-sampling, i.e. takes a subset of random reads from sample(s)
- FilterNonFunctional Filters non-functional clonotypes
- SelectTop Selects a fixed number of top (most abundant) clonotypes from sample(s)
- FilterByFrequency Filters clonotypes based on a specified frequency threshold.
- ApplySampleAsFilter Filters clonotypes that are present in a specified sample from sample(s)
- FilterBySegment Filters clonotypes according to their V/D/J segment
Clonotype表格操作
Clonotype table operations
- PoolSamples Pools clonotypes from several samples together
- JoinSamples Joins a set of samples and generates clonotype abundance profiles
注释
Functional annotation of clonotype tables (antigen specificity, amino acid properties, etc)
- CalcCdrAAProfile Builds a profile of CDR3 regions (V germline, V-D junction, …) using a set of amino-acid physical properties
- Annotate Computes a set of basic (insert size, …) and amino acid physical properties (GRAVY, …) for clonotypes
使用—plot-type png参数获得png格式